
Advances in nucleotide sequencing technologies (deep sequencing) have made it possible in recent years to study the virome – the genomes of all viruses in a host – in human blood, diarrhea, and respiratory secretions; grapevines, and feces of horses and bats. The latter mammals are a particularly important subject because it is known that they harbor the predecessors of several important human viruses, including SARS coronavirus, ebolavirus, marburgvirus, Nipah, Hendra, and rabies viruses. Since there are about 1200 known species of bats, the potential for future human zoonoses is significant.
A multicenter group comprising virologists* and chiropterists (scientists who study bats) has examined the virome of three North American bat species: big brown bats (Eptesicus fuscus), tri-colored bats (Perimyotis subflavus), and little brown myotis (Myotis lucifugus). Deep sequencing was used to analyze fecal and oral samples from 41 bats captured on one night in Western Maryland. The results provide a comprehensive glimpse of the bat virome.
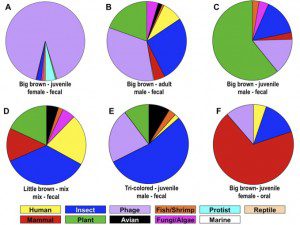
The 576,624 sequence ‘reads€™ (a read is the result of a single sequence reaction, in this study ~250 nucleotides) on six pools of fecal samples revealed an amazing diversity of viral sequences (figure), with representatives of human, other mammals, insect, bacteriophage, fish, shrimp, protist, reptile, plant, avian, fungi, algae, and marine viruses. Among the interesting findings are sequences of three novel coronaviruses from big brown bats. Many coronaviruses have previously been found in bats; the results of this study provide more evidence that these mammals are likely to be sources of future human infections.
Novel viruses of plants and insects were also identified in fecal samples from all bats. This observation can be explained by the bats€™ prodigious appetite for insects, which harbor both insect or plant viruses (the latter transmitted to plants by insect vectors). It seems unlikely that these viruses replicate in bats, but pass through the gastrointestinal tract into the feces.
Perhaps not surprisingly (given the diversity of bacteria that colonize mammalian intestinal tracts), sequences of novel bacteriophages were also identified in fecal samples. One appears to have high sequence identity with a bacteriophage that infects the plague bacterium Yersina pestis. Curiously, such bacteria are not known to colonies animals in the northeastern United States. A second bacteriophage infects strains of E. coli associated with human illnesses. These findings suggest that bats could be involved in the spread of human bacterial pathogens.
The main viral sequences identified in pooled oral samples were of a novel cytomegalovirus. These viruses appear to be common in bats, and have been been detected in many previous studies.
One question that arises from these findings is whether bats are unique in harboring a large collection of diverse viruses. The answer to this question awaits studies of the viromes of other wild animals.
Viral discovery by massive sequencing will no doubt identify many new viruses in a wide range of species. However, this technology cannot answer some of the more intriguing biological questions, such as which hosts support viral replication, and whether it is associated with disease. Answers to these questions will require construction of complete DNA copies of viral genomes and recovery of infectious viruses by transfection of cells in culture.
The title of this post is ‘Bats harbor many viral sequences’, not ‘many viruses’. That’s because no infectious viruses were identified – only parts of their genomes. If you wish to conclude that a certain virus infects bats, you must either isolate the virus in cell culture, or show that the entire viral genome is present in tissues or fluids.
*including Eric F. Donaldson and Matt Frieman, who spoke about this work on TWiV 90 and 65.
Donaldson EF, Haskew AN, Gates JE, Huynh J, Moore CJ, & Frieman MB (2010). Metagenomic Analysis of the Virome of three North American Bat Species: Viral Diversity Between Different Bat Species that Share a Common Habitat. Journal of virology PMID: 20926577

Pingback: Tweets that mention Bats harbor many viral sequences -- Topsy.com