The plaque assay is an essential tool for determining virus titers. The concept is simple: virus infection is restricted to neighboring cells by a semisolid overlay. By counting the number of plaques, the virus titer can be calculated in PFU per ml. A key question is: how many viruses are needed to form a single plaque?
For most animal viruses, one infectious particle is sufficient to initiate infection. This conclusion can be reached by studying the relationship between the number of infectious virus particles and the plaque count. A linear relationship means that one infectious particle can form a plaque. In this case the virus is said to infect cells with one-hit kinetics. This concept is illustrated below. In this figure, the number of plaques produced by a virus with one-hit kinetics or two-hit kinetics is plotted versus the relative concentration of the virus.
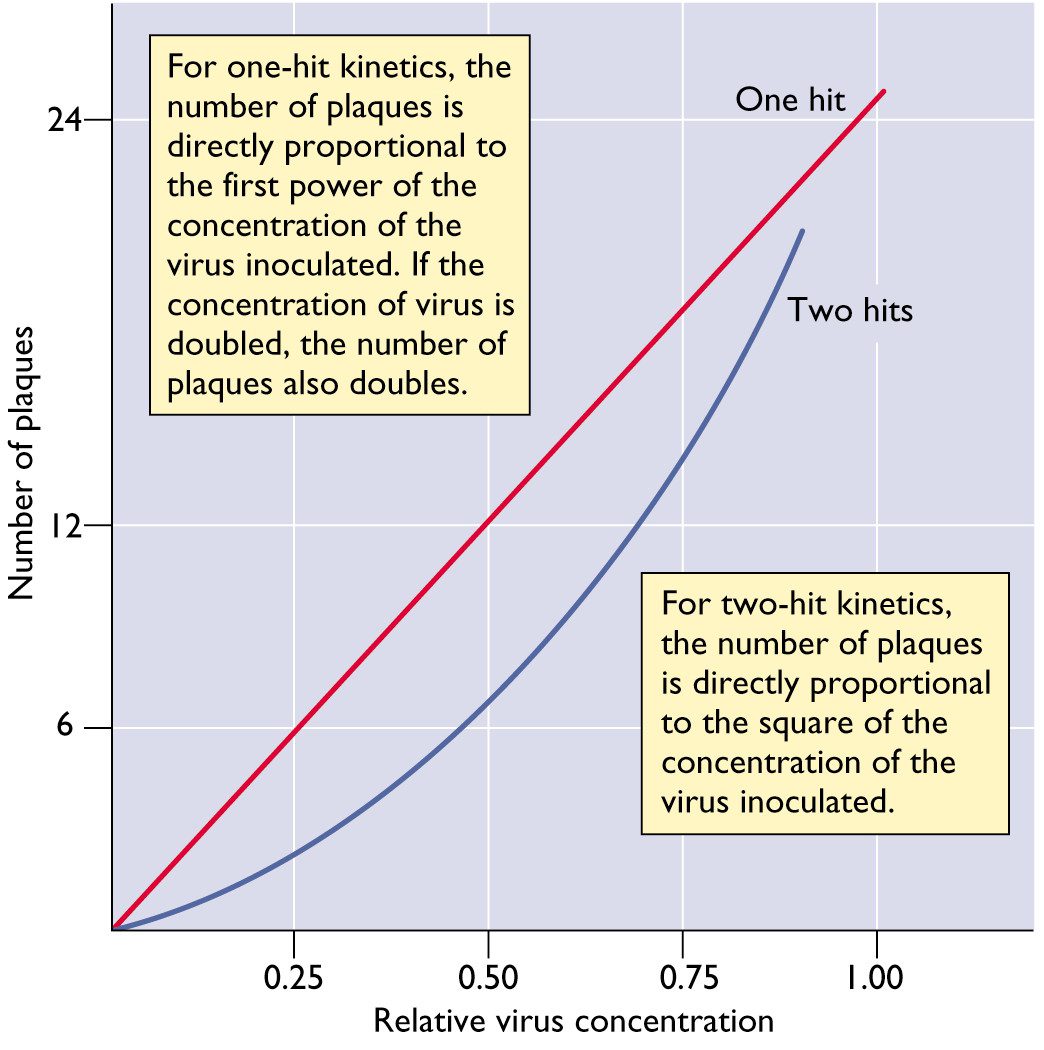

There are some examples of viruses with two-hit kinetics: in other words, two different types of viral particles must infect a cell to initiate the infectious cycle. Examples include the genomes of some (+) strand RNA viruses of plants, which consists of two RNA molecules that are packaged in different particles. The dose-response curve of such viruses is parabolic rather than linear.
When a single virus particle can form a plaque, the viral progeny within the plaque are clones. Virus stocks prepared from a single plaque are called plaque purified virus stocks. To prepare such virus stocks, the tip of a small pipette is inserted into the agar overlay above the plaque. The plug of agar is removed and placed in buffer. The viruses within the agar plug move into the buffer, which can then be used to infect cultured cells. To ensure purity, this process is usually repeated at least one more time. Plaque purification is used extensively in virology to establish clonal virus stocks. The ability to prepare clonal virus stocks was an essential development that permitted genetic analysis of viruses.

Could you explain the effect of defective interfering particles and how they affect the final titer of the stocks? and why by using extremely low MOI increase the efficiency?
Dr. Racaniello, I would like to expand on David Loria's question as well as inquire about potential therapeutic uses for defective interfering particles (DIPs) or genetically modified DIPs. If it is the case that the smaller genomes of DIPs are more efficiently replicated than the full length viral genome, then why is the interference not sufficient to eliminate the viral infection? Is this because of a low probability of coinfection between DIPs and viruses with full length genomes? Along those same lines, in the presence of DIPs, why is there a characteristic alternation from high to low infectious titers produced from passage to passage? By the way, thanks for elevating virology’s standing in social media.
Defective interfering particles arise when virus infections are
carried out serially at high multiplicity of infection. They contain
grossly altered genomes, eg with large deletions, but maintain
essential functions that allow replicating and packaging of genomes.
In co-infections with wild type viruses DI particles show a
replication advantage because they are more efficient at replicating
their shorter genome, and compete with wild type virus for essential
replication proteins. Why they arise at high moi infections is not
known but might be related to their ability to out-compete wild type
virus when replication factors are limiting, as would be the case when
many viruses infect a cell.
Most DI particles require functions provided in trans by the wild-type
virus. In the simplest case, DI particles are unable to produce a
complete viral capsid, and therefore the DI genomes can only be
packaged when a wild-type virus co-infects the same cell. The
oscillation in titers occurs because the DI particles reach
sufficiently high numbers that they interfere substantially with the
'helper' virus. Since the DI particle needs the helper virus, the
result is that DI particle titers decline, until helper virus titers
recover. Hence the oscillating titers of both types of particle.
How many virus particles are typically contained in one plaque?
It would depend upon the particular virus, cell, and the size of the
plaque. From a few thousand to several million.
Prof VRR: Do you have any clue as to why DIP are formed in nature?
I have a clue, but I don't know the real reason – neither does anyone else. They form because they outgrow wild type genomes. When a deletion occurs in the viral genome – possibly by polymerase skipping, which happens randomly – those smaller genomes replicate faster than wild type genomes and eventually interfere with replication of wild type virus.
Hi, Prof. Racaniello,
Thanks for these wonderful posts about influenza virus. I'm studying detection of influenza virus by PCR, and confused by the results. According to my qRT-PCR results, 1 PFU of influenza A H3N2 approximately equals to 100,000 genome copies!! (RNA extracted by Trizol-chloroform, RT with random hexamers, PCR targeting a segment of M gene) I tried samples with concentrations ranging from 1-10,000 PFUs, and the results are quite consistent. And some researchers reported a detection limit of 10-4 TCID50. Then if 1 PFU=0.6-0.7 TCID50=1 viral particle, how come this happens? So how many viral particles does a PFU contains exactly? Thanks.
Hi, Prof. Racaniello,
Thanks for these wonderful posts about influenza virus. I'm studying detection of influenza virus by PCR, and confused by the results. According to my qRT-PCR results, 1 PFU of influenza A H3N2 approximately equals to 100,000 genome copies!! (RNA extracted by Trizol-chloroform, RT with random hexamers, PCR targeting a segment of M gene) I tried samples with concentrations ranging from 1-10,000 PFUs, and the results are quite consistent. And some researchers reported a detection limit of 10-4 TCID50. Then if 1 PFU=0.6-0.7 TCID50=1 viral particle, how come this happens? So how many viral particles does a PFU contains exactly? Thanks.
Pingback: Are all virus particles infectious?
Hi Dr. Racaniello,
Can you please explain the significance of plaque size against plaque numbers.
Â
Which of these indicates viral pathogenesis and replication rate ?
How might PFUs differ from the
number of viruses?