
Both new viruses were isolated by culturing environmental samples in the amoeba Acanthamoeba castellani. Pandoravirus salinus was isolated from shallow marine sediment in a river at the coast of central Chile, and Pandoravirus dulcis was obtained from mud at the bottom of a freshwater pond near Melbourne, Australia. The P. salinus genome is at least 2.77 megabases in length (there is some uncertainty in the actual length due to the presence of repeated sequences at the ends of the DNA), while the P. dulcis genome is 2.47 megabases in length. The smaller P. dulcis genome is a subset of the P. salinus genome.
These new genomes are twice as large as those of previously described viruses, and bigger than the genomes of intracellular bacteria such as Tremblaya (138,927 base pairs) and Rickettsia (1,111,523 bp), some free living bacteria, and many free living Archaea.
While the huge sizes of the Pandoravirus virion and genomes are amazing, I find three other features of these viruses even more remarkable. The first is their atypical replication cycle. The virions are taken into amoebae by phagocytic vacuoles, and upon fusing with the vacuole membrane, the virion contents are released into the cytoplasm via a pore on the virion apex. Within 2-4 hours the cell nucleus is reorganized, and by 8-10 hours new particles appear where the nucleus once was. Pandoravirus DNA and virions are synthesized and assembled simultaneously, in contrast to eukaryotic DNA viruses and phages which fill pre-formed capsids with DNA. Virions are released by 10-15 hours as the cells lyse.
A second amazing feature is that most of the P. salinus open reading frames encode brand-new proteins. Of the 2,556 putative protein coding sequences in the P. salinus genome, 93% have no recognizable counterparts among known proteins. Some of the genes found in large DNA viruses are present, such as those encoding DNA polymerase and DNA-dependent RNA polymerase, and several amino acyl-tRNA synthetases, like members of the Megaviridae. Curiously, many of the Pandoravirus coding regions contain intervening sequences, which must be removed by RNA splicing. This process is known to occur only in the cell nucleus, suggesting that some Pandoravirus transcription occurs in that organelle. The lack of gene homology leads to authors to conclude that ‘no microorganism closely related to P. salinus has ever been sequenced’.
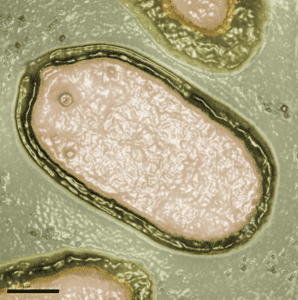
I am also impressed by what the authors describe as the ‘alien morphological features’ of the virions. The oval-shaped particles are 1 micron in length and 0.5 microns in diameter, easily visible by light microscopy. They are wrapped in a three-layered envelope with a pore at one end of the particle, and resemble nothing that has ever been seen before (see photograph).
How much bigger can viruses get? I don’t know the answer but I would guess even bigger than Pandoraviruses. The membranous Pandoravirus particle could easily accommodate even larger genomes. How big can a virus get and still be a virus? The answer to that question is easy: it is a virus as long as it requires a cell for replication.
These remarkable findings further emphasize the need for scientists to pursue their curiosity, and not only work on problems of obvious medical relevance. As the authors write,
This work is a reminder that our census of the microbial diversity is far from comprehensive and that some important clues about the fundamental nature of the relationship between the viral and the cellular world might still lie within unexplored environments.
Continuing their playful naming of giant viruses, the authors note that the name Pandoravirus reflects their ‘lack of similarity with previously described microorganisms and the surprises expected from their future study’.

This is really interesting; since it seems the Pandoraviruses have regions that must be removed by RNA splicing, have they checked to see if those spliced out regions interact further with host Drosha or Dicer (or their equivalents in the amoeba) as they would if they were to function as miRNA’s?
Good question! I’m sure it is one of many things planned. I don’t know of any miRNA studies on these giant viruses as of yet.
“The answer to that question is easy: it is a virus as long as it requires a cell for replication”.
Ummmmm…possibly more correctly, “it is a virus as long as it requires cell components and ribosomes for replication”?
Nice commentary!
Oh, and: “Continuing their playful naming of giant viruses…” – for which they WILL get into trouble with the ICTV!
does pandoravirus cause any disease in human?
You mention “the authors” three times but do not name them or link to the paper(s).
–bks
Oh, nevermind. The picture is linked to the article.
–bks
Is it possible that a virus can evolve out of a degenerated prokaryotic cell, whose genome was deleted enough to make it incapable of replicating on its own, but still can infect living cells?
“They are smaller than genomes of free living bacteria (Mycobacterium, 3,268,203 bp)…”
Larger than some. Haemophilus influenzae is only 1,830,140 bp.
Indeed! It’s bigger than the genomes of some other free living bacteria as well. Fixed.
Not that we know of. But see The largest viral genome from a human for report of isolation of a Megavirus from a human with respiratory disease.
Fixed. I usually include a link but forgot to do it this time.
I’ve always wondered how these giant viruses fare with ICTV. Do they try to get the authors to change, or do nothing?
The ICTV does try to get people to fall in line – and you will find that in the formal Descriptions of Viruses / Virus Taxonomy releases, names that do not meet the criteria for nomenclature are not accepted. As for what happens when they enter the world of popular media, well…B-) Who knows??
Potentially – but these viruses look truly ancient, so if that is what happened, it did so around the dawn of cellular life. It could even be that they evolved as “partner genomes”, which then became independent by picking up a transport function – aka shell or capsid.
This is one of the most competent articles I have read – but why did you keep secret that similar or almost identical organisms were dicovered twice years before in Germany? They had been described in detail without supposing their virus character and have been cited correctly in the article on the novel Pandoravirus by the french authors, i,e, nr 41 and 42 in the refererence list.
Pingback: Pandoravirus, bigger and unlike anything seen before | IB Biology @ SAS
Pingback: TWiV 246: Pandora, pandemics, and privacy
Pingback: TWiV 246: Pandora, pandemics, and privacy | Alan Dove, Ph.D.
I’ve looked at ICTV taxonomy online (http://ictvonline.org/virusTaxonomy.asp?version=2012) and it appears that the Mimivirus family (with species Acanthamoeba polyphaga mimivirus) and the Cafeteriavirus family (with species Cafeteria roenbergensis virus) have all been accepted as official names. So maybe ICTV is not so rigid after all.
Mutter grumble what IS the world coming to mutter mutter…B-)
Actually, see also here: http://www.virologyj.com/content/10/1/254/abstract
It really probably is time for a rethink of what we name our favourite organisms, and why.
Pingback: Pandoravirus, bigger and unlike anything seen before | Viral Bioinformatics Resource Center
Pingback: Monster Viruses – Mimivirus and Pandoravirus (Comparative Understanding) | Biomysteries
Pingback: Possibility of a Missing Link Between Viruses and Living Organisms | David Kish
Pingback: TWiV 246: Pandora, pandemics, and privacy | This Week in Virology
does anyone know if the pandoravirus has any receptors to bind to the vocule?
Pingback: How viruses infect a cell within a cell
Pingback: parasitology | [Veterinary and Medical Sciences
Pingback: How viruses infect a cell within a cell – Virology