
The first genome sequences reported were from the initial three H7N9 isolates: A/Shanghai/1/2013, A/Shanghai/2/2013, and A/Anhui/1/2013. These were followed by genome sequences from A/Hongzhou/1/2013 (from a male patient), A/pigeon/Shanghai/S1069/2013), A/chicken/Shanghai/S1053/2013), and A/environment/Shanghai/S1088/2013, the latter three from a Shanghai market.
Analysis of the viral genome sequences reveals that all 8 RNA segments of influenza A/Shanghai/1/2013 virus are phylogenetically distinct from A/Anhui/1/2013 and A/Shanghai/2/2013, suggesting that the virus passed from an animal into humans at least twice. Similar viruses have been isolated from pigeons and chickens, but the source of the human infections is not known. There is as yet no evidence for human to human transmission of the H7N9 viruses, and it seems likely that all of the human infections are zoonotic – transmission of animal viruses to humans. Since the H7N9 viruses are of low pathogenicity in poultry, infected animals may not display disease symptoms, further facilitating transmission to humans.
The RNA sequences reveal that the H7N9 viruses isolated from humans are all triple reassortants, which means that they contain RNA segments derived from three parental viruses. The gene encoding the hemagglutinin protein (HA) is most closely related to the HA from A/duck/Zhejiang/12/2011 (H7N3), while the NA gene is most similar to the NA gene from A/wild bird/Korea/A14/2011 (H7N9). The remaining 6 RNA segments are most related to genes from A/brambling/Beijing/16/2012-like viruses (H9N2). The type of animal(s) in which the mixed infections took place is unknown.
Some observations on the relatedness of these sequences:
- A/Shanghai/2/2013, A/Anhui/1/2013, and A/Hangzhou/1/2013 were isolated in distant cities yet have over 99% identity. The pigeon, chicken, and environmental isolates are also very similar except for one gene of A/pigeon/Shanghai/S1069/2013. Long-range shipping of infected poultry might explain these similarities.
- There are 53 nucleotide differences between A/Shanghai/1/2013 and A/Shanghai/2/2013. Perhaps A/Shanghai/1/2013 and the remaining viruses originated from different sources.
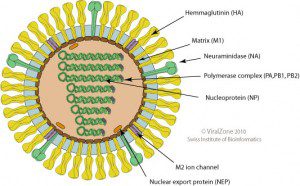
When the gene sequences of these human viral isolates are compared with closely related avian strains, numerous differences are revealed. The locations of the proteins in the influenza virion are shown on the diagram; click for a larger version (figure credit: ViralZone).
- All seven H7N9 viruses do not have multiple basic amino acids at the HA cleavage site. The presence of a basic peptide in this location allows the viral HA to be cleaved by proteases that are present in most cells, enabling the virus to replicate in many organs. Without this basic peptide, the HA is cleaved only by proteases present in the respiratory tract, limiting replication to that site. This is one reason why the H7N9 viruses have low pathogenicity in poultry.
- All seven viruses have a change at HA amino acid 226 (Q226L) which could improve binding of the viruses to alpha-2,6 sialic receptors, which are found throughout the human respiratory tract. Avian influenza viruses prefer to bind to alpha-2,3 sialic acid receptors. This observation suggests that the H7N9 isolates should be able to infect the human upper respiratory tract (alpha-2,3 sialic acid receptors are mainly located in the lower tract of humans). However, viruses which bind better to alpha-2,3 sialic acids still bind to alpha-2,6 receptors and can infect humans.
- All seven viruses have a change at HA amino acid 160 from threonine to alanine (T160A). This change, which has been identified in other circulating H7N9 viruses, prevents attachment of a sugar to the HA protein and could lead to better recognition of human (alpha-2,6 sialic acid) receptors.
- Five amino acids are deleted from the neuraminidase (NA), the second viral glycoprotein, in all seven viruses. In avian H5N1 influenza virus this change may influence tropism for the respiratory tract and enhance viral replication, and might regulate transmission in domestic poultry. This change is believed to be selected upon viral replication in terrestrial birds.
- One of the viruses (A/Shanghai/1/2013) has an amino acid change in the NA glycoprotein associated with oseltamivir resistance (R294K).
- An amino acid change in the PB1 gene, I368V, is known to confer aerosol transmission to H5N1 virus in ferrets.
- An amino acid change in the PB2 gene, E627K, is associated with increased virulence in mice, higher replication of avian influenza viruses in mammals, and respiratory droplet transmission in ferrets.
- Changes of P42S in NS1 protein, and N30D and T215A in M1 are associated with increased virulence in mice, but these changes are also observed in circulating avian viruses.
- All seven viruses have an amino acid change in the M2 protein known to confer resistance to the antiviral drug amantadine.
- All seven viruses lack a C-terminal PDZ domain-binding motif which may reduce the virulence of these viruses in mammals.
For the most part we do not know the significance of any of the amino acid changes for viral replication and virulence in humans.
I believe that these H7N9 viruses might take one of two pathways. If they are widespread in birds, they could spread globally and cause sporadic zoonotic infections, as does avian influenza H5N1 virus. Alternatively, the H7N9 viruses could cause a pandemic. Influenza H7N9 virus infections have not occurred before in humans, so nearly everyone on the planet is likely susceptible to infection. Global spread of the virus would require human to human transmission, which has not been observed so far. Some human to human transmission of avian H7N7 influenza viruses was observed during an outbreak in 2003 in the Netherlands, but those viruses were different from the ones isolated recently in China. Whether or not these viruses will acquire the ability to transmit among humans by aerosol is unknown and cannot be predicted. If a variant of H7N9 virus that can spread among humans arises during replication in birds or humans, it might not have a chance encounter with a human, or if it did, it might not have the fitness to spread extensively.
What also tempers my concern about these H7N9 viruses is the fact that the last influenza pandemic (H1N1 virus) took place in 2009. No influenza pandemics in modern history are known to have taken place 4 years apart, although only 11 years separated the 1957 (H2N2) and 1968 (H3N2) pandemics. I suppose that is not much consolation, as there are always exceptions, especially when it comes to viruses.
Meanwhile a vaccine against this H7N9 strain is being prepared (it will be months before it is ready), surveillance for the virus continues in China and elsewhere, and health agencies ready for a more extensive outbreak. These are not objectionable courses of action. But should this be our response to every zoonotic influenza virus infection of less than 100 cases?
Sources
Human Infection with a Novel Avian-Origin Influenza A (H7N9) Virus.

Yep, never can be too careful. Let’s get extremely hysterical during the first months. Say, “This is the end of everything!!!!” Run around. Scream. Get naked. And, then when goes away or turns out to not be all that very deadly, people will relax and accept that, you know, the birds and the pigs have always been plotting to kill us.
Prof. Racaniello, thank you for the comprehensive genetic analysis of H7N9 virus. I think the amino acid change that is known to confer resistance to antiviral drug amantadine is in the M2 ion channel protein and not in M1 protein.
I find it curious that the parental flu strains are as widely separated as they are. Would the likely mixing bowl have been avian or some other animal such as a pig.
The Human race has chosen to feed on pigs and poultry. Production has increased by 300 and 700% respectively, over the last 40 years and the trend continues upwards. Many zoonotic incidences like this are to come and we will eventually experience a fatigue of hystery. But the question remains: can we really do anything to prevent this from happening?
Based on observations from the field and my own experiments working on reassortment (read: I have no data to back this up) I would guess the first reassortment that paired the HA and NA was probably in ducks. That virus then transmitted to chickens and reassorted with an H9N2 that was already there.
Yes of course, thanks for pointing that out. Fixed. Good to have so many error checking readers!
I don’t know the answer, but open meat markets are probably not helpful. Infections will still occur in domestic swine and poultry, but at least transmission to humans might be somewhat restricted.
I think with our advancements in science, medicine, and technology (i.e. RNA vaccines, antiviral treatment, realtime global communication), robust response and survalliance should be the standard response for novel viral human-zooinotic outbreaks. As we become more efficient at responding to such events cost and psycological stress should be mitigated.
Pingback: Virologists plan influenza H7N9 gain of function experiments
Pingback: নতà§à¦¨ নতà§à¦¨ ফà§à¦²à§ à¦à¦¾à¦‡à¦°à¦¾à¦¸à§‡à¦° আগমন কি বিবরà§à¦¤à¦¨à§‡à¦° উদাহর
Pingback: নতà§à¦¨ নতà§à¦¨ ফà§à¦²à§ à¦à¦¾à¦‡à¦°à¦¾à¦¸à§‡à¦° আগমন কি বিবরà§à¦¤à¦¨à§‡à¦° উদাহর