As we discussed previously, attachment of all influenza A virus strains to cells requires sialic acids. However, there are a number of chemically different forms of sialic acids, and influenza virus strains vary in their affinity for them. These differences may determine which animal species can be infected.
In the example shown below, sialic acid is linked to the sugar galactose by what is called an alpha(2,3) linkage. This means that the carbon atom at position number 2 of the sialic acid hexose is joined, via an oxygen atom, to the carbon at position 3 of the hexose of galactose.
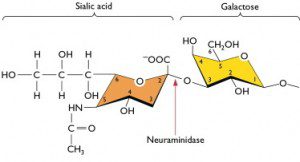

Avian influenza virus strains preferentially bind to sialic acids attached to galactose via an alpha(2,3) linkage. This is the major sialic acid on epithelial cells of the duck gut. In contrast, human influenza virus strains preferentially attach to sialic acids attached to galactose by an alpha(2,6) linkage. This is the major type of sialic acid present on human respiratory epithelial cells. Alpha(2,3) linked sialic acids are found on ciliated epithelial cells, which are a minor population within the human respiratory tract, and also on some epithelial cells in the lower tract.
This receptor specificity has implications for human infection with avian influenza virus strains. For example, highly pathogenic avian H5N1 influenza viruses undergo limited replication in the human respiratory tract due to the presence of some cells with alpha(2,3) linked sialic acids. However, efficient human to human transmission requires that the avian viruses recognize sialic acids with alpha(2,6) linkages. Consistent with this hypothesis, the results of studies of early influenza virus isolates from the 1918, 1957, and 1968 pandemics suggest that these viruses preferentially recognized alpha(2,6) linked sialic acids.
Epithelial cells of the pig trachea produce both alpha(2,3) and alpha(2,6) linked sialic acids. This is believed to be the reason why pigs can be infected with both avian and human influenza virus strains and serve as a ‘mixing vessel’ for the emergence of new viruses. However, Peter Palese has his doubts, as recorded in The Great Influenza by John Barry:
Dr. Peter Palese….considers the mixing-bowl theory unnecessary to explain antigen shift: “It’s equally likely that co-infection of avian and human virus in a human in one cell in the lung [gives] rise to the virus…There’s no reason why mixing couldn’t occur in the lung, whether in pig or man. It’s not absolute that there are no sialic acid receptors of those types in other species. It’s not absolute that that avian receptor is really that different from the human, and, with one single amino acid change, the virus can go much better in another host”.
Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, & Kawaoka Y (1998). Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. Journal of Virology, 72, 7367-7373
Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, & Kawaoka Y. (2000). Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. Journal of Virology, 74, 8502-8512

Relevant post on this topic over at Effect Measure: http://scienceblogs.com/effectmeasure/2008/01/b…
Professor Racaniello,
I am very much enjoying your blog. Your enthusiasm for virology is truly infectious. (Couldn't resist.)
It is amazing how much you and your fellow virologists have managed to discover about these little beasts–all to our great benefit. Thank you very much.
I'm glad you are enjoying virology blog. The new strain of influenza
has brought many new readers who are interested in learning about
viruses. I've been teaching virology for 30 years, but I've taught
more people in the past week than in all those years.
Well, you're doing a fine job, and I hope you keep it up. Now that I've found your blog, I intend to keep it on my reader. 🙂
I wanted to say that I found the series of posts describing the different aspects (structure of influenza virions and how the viral RNAs can encode one or more proteins) and steps on the virus life cycle (attachment and entry of the virion, translation of mRNA into protein, genome replication, assembly of new particles, and release of particles from the cell) incredibly helpful, specially since these can be easily translated into the general case. By the way, great illustrations (made by you?).
I'm looking forward to read the following posts on the subject!
OK, I'm a little confused with the mixing issue. One type of cell has (2,3) linked, another type of cell has (2,6). Pigs have a lot of both; humans have only a few (2,3). Good so far.
But how does mixing occur unless a single cell expresses both (2,3,) and (2,6)? I can see this phenotypic mixing if a single cell is co-infected, and Gallo showed this decades ago. But merely co-infecting the whole animal doesn't make the case because the two types of virus will be internalized by two different cells.
Pingback: The D225G change in 2009 H1N1 influenza virus is not a concern
Pingback: The D225G change in 2009 H1N1 influenza virus is not a concern « FluTrackers
Can any one let me know what is the sequence of main epitops on H1 .I am preparing polyclonal antibody against this protein .
Good question. Epithelial cells of the pig trachea have both (2,3) and
(2,6) linkages on the same cells. This has been shown by staining
sections of pig trachea with lectins (sugar-binding proteins) specific
for either (2,3) or (2,6) sialic acids. The cells react with both
lectins.
Sequences of influenza H1 antigenic sites can be found in Science.
2004 Mar 19;303(5665):1866-70. Structure of the uncleaved human H1
hemagglutinin from the extinct 1918 influenza virus.
Can any one let me know what is the sequence of main epitops on H1 .I am preparing polyclonal antibody against this protein .
Good question. Epithelial cells of the pig trachea have both (2,3) and
(2,6) linkages on the same cells. This has been shown by staining
sections of pig trachea with lectins (sugar-binding proteins) specific
for either (2,3) or (2,6) sialic acids. The cells react with both
lectins.
Sequences of influenza H1 antigenic sites can be found in Science.
2004 Mar 19;303(5665):1866-70. Structure of the uncleaved human H1
hemagglutinin from the extinct 1918 influenza virus.
i had kept the questions for a week long and couldnt find it out. but finally, get the answer here. I love it. Cheers~!!!
any one have any idea about which reported strains replicate in or infects alpha-2,3 SA receptor bearing cells? how about for alpha-2,6 expressing cells?
can I ask of where sialic acid attaches to in a human skin mast cell protein?Â
Sialic acid is the last sugar moiety in the chain of sugars attached
to a glycoprotein.
can you plz provide me the activated structure of sialic acid and NA during the reaction.
Javed Hussain
If influenza binds to the terminal sialic acid residue at a site distant from the cell membrane, how is this “sensed” by the cell, leading to receptor-mediated endocytosis? Does binding cause a conformational change in the protein to which the sialic acid is attached?
where and how influenza virus strains are binding to the sialic acid attached to galactose (2,6) linkage in humans ?
Pingback: Avian H5N1 influenza and biosecurity on Science Friday
Pingback: Influenza H5N1 is not lethal in ferrets after airborne transmission
Pingback: First human infections with avian influenza H7N9 virus
Pingback: A single amino acid change switches avian influenza H5N1 and H7N9 viruses to human receptors
If the affinity of HA for sialic acid is low, how can efficient attachment occur with the low affinity interaction?
Because there are multiple HA-sialic interactions, not just one, as the virion binds to the cell surface. Multiple low-affinity interactions lead to a virus-cell interaction of high avidity. This is typical of virus-receptor interactions.
Hello Professor,
Can I use some of your images in my presentation for an in-class assignment? How do I cite your work?
Thank you,
Pingback: Influenza A viruses in bats
Pingback: Changing influenza virus neuraminidase into a receptor binding protein
Pingback: Yet another avian influenza virus, H10N8, infects humans
Hello professor
Do you think ssDNA viruses evolve eventually similar to ssRNA viruses but adapt to their environment quicker then ssRNA.
Hello Professor,
Thank you for your blog. I have one question about the sialic acid receptors. My understanding is that the avian influenza virus has a preferential binding to alpha(2,3) linked sialic acids, but when it becomes pandemic, in order to transmit from human to human, it can also bind to alpha(2,6) linked sialic acids. Therefore the binding property changes. Is there any human influenza virus that can infect avian? My point is can human influenza virus prefer to bind alpha(2,3) linked sialic acids?
how colorimetric detection of sialic acid attached to the influenza virus can be done??
v simple and understandable good job
can u plzzz hlp me prof i need some articles on human influenza plzzz recent articles
scholar.google.com — this is the best site for looking for academic, peer-reviewed papers.
Use it, and type in whatever you’re looking for, for influenza. “Influenza biochemistry”, “Influenza enzymes”, etc. Then you can use the advanced search to limit what years you want papers from, or even what journals you look through.
http://www.nature.com/nrmicro/journal/v13/n1/abs/nrmicro3405.html
Here’s a review article about gain-of-function mutations with influenza that came out not long ago!
i need human influenza A sequences of research performed in pakistan plzzz if u can hlp mei have an article but the accesion number given in that article is un reachable means show no result
and thank u for the article but its unreachable 🙁
we have to buy it
can any body tells me that we have other sialic acid receptor too in our body why not influenza virus attach to them ?? and i am still confuse about their other fucntion in body