
A Q-beta phage population is in a dynamic equilibrium with viral mutants arising at a high rate on the one hand, and being strongly selected against on the other. The genome of Q-beta cannot be described as a defined unique structure, but rather as a weighted average of a large number of different individual sequences.
This variation, which is a consequence of the error prone nature of viral replication, has since been confirmed using other viral systems. Virologists now understand that virus populations are not made of a single member with a defined nucleic acid sequence. Rather, they are dynamic distributions of nonidentical but related members called a quasispecies. It was given this name because the classical definition of species – an interbreeding population of individuals – has little meaning for viruses.
The consequence of a quasispecies is that most viral infections are initiated not by a single virion, but a population of particles. The progeny produced after this infection results from selective forces that operate inside the infected host. The virions that go on to infect a new host have passed through another set of external selective forces. A steady-state population of a viral quasispecies consists of a vast number of particles.
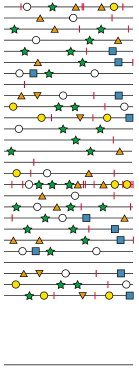
The diagram above shows a small subset of the viral genomes that are present in a virus stock. Genomes are indicated by lines, and mutations are shown by different symbols. The consensus sequence for this population is shown as a line at the bottom. There are no mutations in the consensus sequence, even though every viral genome contains mutations. This is because no mutation is present at sufficiently high levels to achieve a consensus at any position.
Although this concept may seem abstract, as we consider more aspects of viral biology it will become obvious. A key point is that the genome sequences of viruses cluster around an average sequence, but every genome is probably different from that consensus. This means that sequences of the new influenza H1N1 viruses on NCBI or GISAID sequences represent a consensus, and do not represent the population that infected any of the thousands of individuals in Mexico during the past month. Luis Villareal, a prominent evolutionary virologist, had the following thoughts about quasispecies and the current influenza epidemic:
Flu researchers believe in the master template as being the fittest type. I think this has been to their detriment and is currently confusing the field as we witness the evolution of emergence. I would be willing to bet money, that if the quasispecies composition were measured, we would see clear differences between those patients that died in Mexico compared to the much less virulent outcome in the USA.
Until recently it was not possible to know the sequences of all the viral genomes present in a population such as that illustrated in the figure. The development of deep sequencing methods such as 454 pyrosequencing has now made it possible to study the quasispecies. This method can detect the individual variants within a viral population.
Tomorrow we’ll consider how selection pressures within a host can change the quasispecies.
Domingo, E. (1978). Nucleotide sequence heterogeneity of an RNA phage population Cell, 13 (4), 735-744 DOI: 10.1016/0092-8674(78)90223-4

I'm very much enjoying this series, thanks.
You write: “consequence of a quasispecies is that most viral infections are initiated not by a single virion, but a population of particles”
I'm confused. Presumably the population diversity is very high in an infected animal. Presumably when the infection is passed from one animal only some sample of that population moves over into the next animal. The quote seems to be saying something about how complete a sample is usually passed on. Of course the story book version of viral infection involves a single infective viral particle doing the job. If I'm reading that quoted bit right your saying that in most cases significant representative sample of the entire quasispecies moves onto the next victim.
Infections of animals are never initiated by a single particle; almost
always by many (I shouldn't say never, it always comes back to get me,
but it's likely extremely rare). A single particle, even a few hundred
or thousand, are not enough – viral infection is quite inefficient in
an animal host. Probably millions of particles are needed, but it
depends on the virus and host. So your statement below is absolutely
correct: “in most cases significant representative sample of the
entire quasispecies moves on to the next victim”.
Facinating! It tastes like a 'sum over histories' interpretatioin of quantum mechanics where the wave functions for all the probablities are present, and some cancel each other out and what is left is the 'consensus' reality that is determined by the observer (who is the 'host'
I'm strictly an amateur scientist, so you don't need to pay much attention to my musings. But, it facinates me anyway. Thanks for the article.
That's just so cool. There is a tempting analogy to be made. It's as if that population (and it's statistics) are the infectious agent, that agent the individual representative of the quasispecies – analagous to how the individual animal is a representative of his species. The entire population is then what evolves. And it's statistics are a work around for the errors at the level of reproducing a single particle. But the analogy looks more fun or tempting than useful.
Thanks!
I've never understood what quantitative difference separates prokaryotic and eukarytoic species from viral quasispecies, given that most prokaryotic/eukaryotic species also exist as a spectrum of mutants ie. are polymorphic. Also, why are RNA viruses regarded as quasispecies, but not DNA viruses?
Pingback: Viral quasispecies and bottlenecks
Is it therefore fair to describe the “error-prone” ways of RNA synthesis as the “variation-prone ways of RNA synthesis?
I'm not a mathematician, but you can imagine that quasispecies,
dealing with population biology, has its share of mathematical
analyses.
You are absolutely correct – viral populations, not individual
mutants, are the target of selection. As you will see, if you limit
diversity, the population suffers.
Variation would be a fair way to describe it; just remember that some
variations are neutral, some are lethal, and some advantageous.
I'm wondering if you would use Wiener's statistical mechanics to describe it. Seems likely.
http://en.wikipedia.org/wiki/Statistical_mechanics
Oh oh. Looks like I better brush up on my eigenvectors!
(Page 88)
http://tinyurl.com/pteeq4
Wow, that's some book. By the head of the Department of Theoretical
Biology, Institute for Advanced Study, Princeton. Some smart people
there. I'm talking Friday on TWiV with a former scientist from that
Institute, on the new flu sequences.
Thanks, I'm glad you like it! My doctorate is in another field so I was trying to find a handy but very competent reference and that seemed to do the trick for me. It's way near the top of my bookmarks.
Any biological population is a mix of sequences and therefore a quasispecies. RNA viruses get all the attention because they cannot correct errors and their sheer numbers makes their quasispecies more extensive than any other.
Would this be the reason for some viruses having so many serotypes? (classic one I have in mind is rhinovirus)
Yes, such variation would indeed explain viruses like rhinovirus that
exist as many serotypes. But much more variation occurs than is
evident by serotype. For example, there are about 100 serotypes of
rhinovirus, but the virus continues to undergo variation that does not
seem to produce new serotypes.
Yes, such variation would indeed explain viruses like rhinovirus that
exist as many serotypes. But much more variation occurs than is
evident by serotype. For example, there are about 100 serotypes of
rhinovirus, but the virus continues to undergo variation that does not
seem to produce new serotypes.
Pingback: Influenza variations | Mystery Rays from Outer Space
yes i agree completely we can never make viruses fall under taxonomical specieses as they are constantly changing…”quasispecies” is an apt terminology
Thank you for your fascinating and beneficial short classes,
Does mutation occur randomly or there are some hot-spot regions? I mean, are there any conserved regions or divergent regions in RNA virus genomes? Thanks again…
Looks like you’ve done your research very well.
Pingback: Describing a viral quasispecies
if i may. DNA viruses actually replicate with a mutation rate of the same order of magnitud than RNA viruses. Even when they are replicated entirely by the cell machinary, they achieve genetically heterogeneous populations. Many other mechanisms (additionally to error-prone polymerases) has been proposed.
Pingback: Viral variation in single cells
Stumbled across this blog and was wondering are these comments so long ago. I am a patient myself on dialysis and have been infected with multiple genotypes(1b, 3, 6) of hepatitis c and looking for answers if I have been having patient to patient infection and the dialysis unit trying to hide. Anybody willing to help?
Pingback: Good viruses visiting bad neighborhoods
Pingback: Good viruses visiting bad neighborhoods - Virology
Pingback: Good viruses visiting bad neighborhoods - VETMEDICS