
In the old days, solving a virus structure was a big deal. A virus is, after all, a very large assembly of many proteins. To solve the structure of a virus – which will tell us the location of the amino acid chains in three dimensional space – was a technical tour de force. It was necessary to purify large amounts of virus particles, and then find the conditions to produce crystals, a hit and miss affair. If you were lucky to grow virus crystals – which could take a year or more – you then crossed your fingers to see if they diffracted in an X-ray beam. When X-rays are aimed at a crystal, the beams bounce off atoms in the crystals, and their reflections provide information on where the atoms are located. Finally you could collect the diffraction data, do a lot of math on a computer, and determine the three dimensional structure.
The first virus structure to be solved by X-ray crystallography was of a plant virus, tomato bushy stunt virus in 1976, followed by poliovirus and rhinovirus in 1985. Many X-ray structures of viruses have been solved, with resolutions less than 2 Angstroms that allow us to see not only the amino acid chain, but all the atoms in the side chains.
The Zika virus structure was not solved by X-ray crystallography. It was done by cryo-electron microscopy (cryo-EM) and image reconstruction. It’s easier and faster than X-ray crystallography, and can achieve comparable resolutions.
It is not necessary to produce crystals to determine structures by cryo-EM. Instead, samples of purified viruses are rapidly frozen and photographed with an electron microscope at very low temperatures. This procedure preserves native structure, and allows visualization of the contrast inherent in the virus particle. Photographs of thousands of virus particles – each in a slightly different orientation – are taken and processing computationally to create the final three-dimensional image.
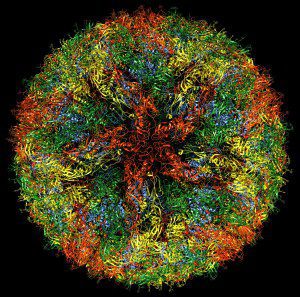
The cryo-EM structure of Zika virus tells us how the virus particle is put together. It looks very much like other flaviviruses, which consist of a membrane surrounding the capsid, which in turn carries the viral RNA genome. Inserted into the membrane are 180 copies of the viral proteins E and M. Although inserted in a fluid lipid bilayer, they are arranged with a symmetry that reflects their contacts with the underlying icosahedral capsid. In the illustration, which I produced from the freely available cryo-EM data, you can clearly see five copies of the E glycoprotein (red) at one five-fold axis of symmetry.
One structural difference between Zika virus and other flaviviruses is a loop of amino acids exposed on the surface of the particle. This sequence of the E glycoprotein, and a sugar molecule attached to it, might be involved in regulating Zika virus tropism and pathogenesis. The ability of West Nile virus to enter the central nervous system of mice has been linked to glycosylation at a similar position, while cell receptors are thought to attach to sugars on the dengue virus capsid.
The authors of the Zika virus cryp-EM structure have produced an animation which illustrates aspects of the structure (below). Watch my lecture on virus structure for more information how viruses are put together.
Updated 7 April 2016 to provide an explanation of how the sugar attached to the E glycoprotein of Zika virus might regulate tropism and pathogenesis.

Pingback: WHO’s Afraid of Little Zika? | al fin next level
yeah boi
yeah boii