by Gertrud U. Rey

Satellite viruses lack one or more of the elements needed to replicate, and thus they depend on co-infection with a helper virus that can provide the missing components. In a well-known example of a satellite-helper system, Hepatitis D virus requires the presence of its helper Hepatitis B virus to make copies of itself. Until recently, we were unaware of the existence of satellite viruses of bacteriophages – viruses that infect bacteria.
A new study by Tagide deCarvalho and colleagues has revealed two satellite-helper systems in bacteriophages that infect Streptomyces bacteria. The bacteriophages (“phages” for short) were identified under the Science Education Alliance – Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program, which allows students to sample soil and identify and name novel phages using a combination of microbiological and computational biology techniques.
Phages isolated from the environment can be detected and quantified using a plaque assay, a technique that involves addition of purified samples (e.g., from soil) to agar plates containing bacteria. If the bacteria are susceptible to infection with phages from a particular sample, the phages will infect and kill the bacterial cells and produce clear spots or “plaques” in those locations on the agar. The phages can then be isolated from the plaques, purified, and their genomes can be sequenced to determine their identity.
The authors of the study noticed that purified isolates from soil samples thought to contain two newly identified phages “Flayer” and “Mulch” each produced two different types of plaques, suggesting that Flayer and Mulch actually each consisted of two different types of phages. Consistent with this observation, sequencing of the DNA extracted from the plaques revealed two different sequences for each of Flayer and Mulch isolates – a long sequence, presumably from a bigger phage, and a much shorter one, presumably from a smaller phage. However, when phages isolated from the plaques were used to re-infect new bacteria, only the ones with the longer sequences produced plaques, suggesting that only the bigger phage from each of the Flayer and Mulch isolates was infectious on its own. Based on these observations, the authors tentatively concluded that Flayer and Mulch were two individual satellite-helper systems with the smaller phages (i.e., the satellites) being dependent on the bigger ones (i.e., the helpers) for infection. The authors also noted that the short sequences were much more abundant than the long ones, suggesting that replication of satellite viruses using helper elements occurs at the expense of replication of the helpers. For ease of reference, the helpers were thereafter named MindFlayer and MulchMansion and their satellites were named MiniFlayer and MulchRoom, respectively.
The genomic material of phages is enclosed in a capsid head that is connected to a short collar and a long tail, which facilitates attachment and injection of genomic DNA into host bacterial cells. Phages are classified as virulent or temperate based on their replication cycles. Virulent phages actively replicate and lyse the host cell once their replication cycle is complete. Temperate phages either integrate their genomes into the host genome or linger in the host cell cytoplasm as circular double-stranded DNA “plasmids” until replication conditions become favorable. Satellite viruses typically infect a cell and stick around using temperate phage mechanisms until the cell is infected by a helper virus. The authors discovered that MulchRoom encodes genes associated with temperate phage properties, suggesting that it can remain in the host cell until MulchMansion infects the cell at a later time. In contrast, MiniFlayer does not encode any such genes, suggesting that it is likely a virulent phage that cannot integrate into the host genome or form a plasmid inside the host cell. This finding implies that MiniFlayer requires MindFlayer for cell entry and must presumably wait outside a prospective host cell until MindFlayer shows up. However, such an assumption raises the question of how MiniFlayer synchronizes its timing of infection with that of MindFlayer.
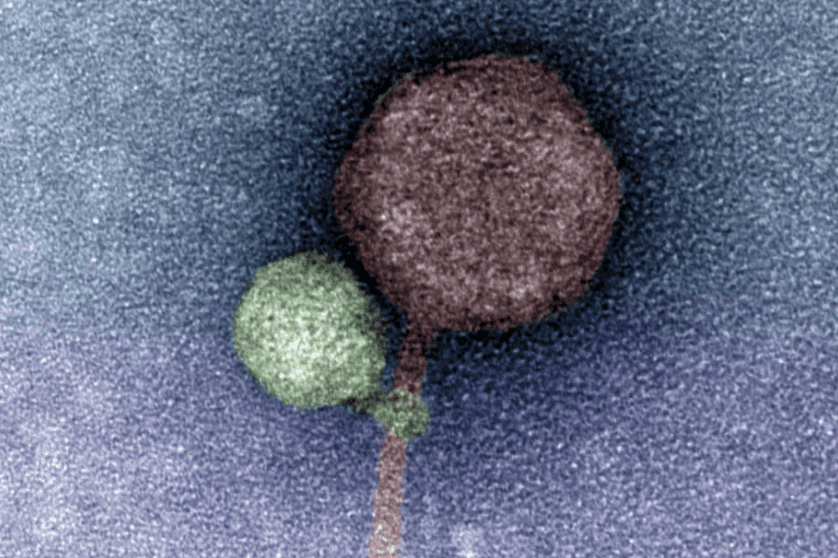
Perhaps the most intriguing discovery in this study was made by electron microscopy, with an abundance of images showing the tail of MiniFlayer apparently wrapped around MindFlayer’s collar and/or tail (imaged). Although the purpose of this interaction is unclear, it is possible that the close association of the virus tails during attachment to a host cell mediates the transfer of both viral genomes into the host cell. If this is true, the exact mechanism by which this transfer occurs still needs to be defined.
Phage satellites make an interesting addition to the evolutionary tango between phages and their hosts. I look forward to learning more about how phages and other viruses interact with each other as imaging technologies and other molecular techniques improve.
[This paper was discussed in detail on TWiV 1067. MiniFlayer has also been repeatedly dubbed a “vampire virus” in the popular press – see for example this article in the Washington Post. The material in this blog post is also covered in Catch This Episode 51.]

“the authors tentatively concluded” … it could also be that the host bacteria which replicated the large phage had inherent protection of some sort inhibiting the smaller satellite phage.