We’ve briefly considered the structure of influenza virions and how the viral RNAs can encode one or more proteins. Now we’ll consider how influenza viruses multiply.
Viruses are obligate intracellular parasites: they cannot reproduce outside of a cell. The production of new infectious particles must take place within a cell. Upon entering cells, viruses parasitize the host machinery to produce new viral progeny. The sum total of all the events that take place in a virus-infected cell is called the infectious cycle, or viral replication. Virologists artificially divide the infectious cycle into steps to make it easier to study. The steps include attachment and entry of the virion, translation of mRNA into protein, genome replication (producing more RNA or DNA), assembly of new particles, and release of particles from the cell. We’ll consider each of these steps, and then move on to a discussion of how influenza virus infects us and causes disease.
Today we’ll focus on the first step, attachment of the virion to cells. Here is a typical cell. I’m sure everyone is familiar with it, but it doesn’t hurt to review.
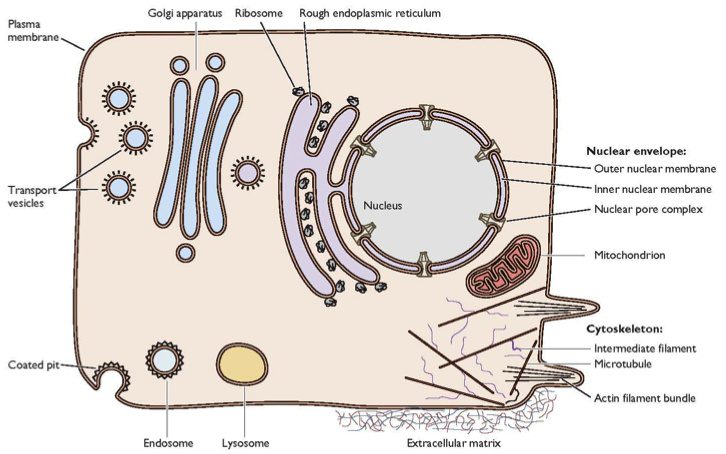

You can see that there is a substantial barrier to anything getting into this cell – the plasma membrane. Viruses have evolved different ways to get around this. But what they all have in common is that virions must first attach to a receptor on the plasma membrane in order to enter the cell. Every virus has a specific receptor that it attaches to, and in turn there is a particular viral protein that binds this cell receptor. Here is an illustration of an influenza virion binding to its cell receptor.
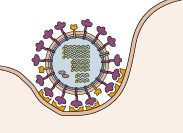

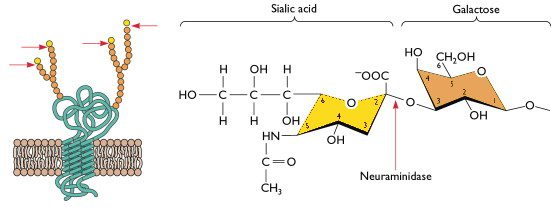
You can see the individual ‘spikes’ on the virion binding to a structure on the cell. The influenza viral spike that attaches to the cell receptor is the HA protein – hemagglutinin. The cell receptor is sialic acid – a small sugar that is attached to many different proteins on the cell surface. Here’s what sialic acid looks like.
On the left is a drawing of a cell protein embedded in the plasma membrane. The interior of the cell – cytoplasm – is at the bottom. Part of the protein crosses the membrane, and there are also parts on the cytoplasmic and extracellular sides. The spheres are sugars that are attached to many proteins (protein + sugar = glycoprotein). Sialic acid is always the last sugar in a chain that is attached to a protein. On the right is the chemical structure of sialic acid; the next sugar, to the right, is galactose. Influenza virions attach to cells when the HA grabs onto the very small sialic acid.
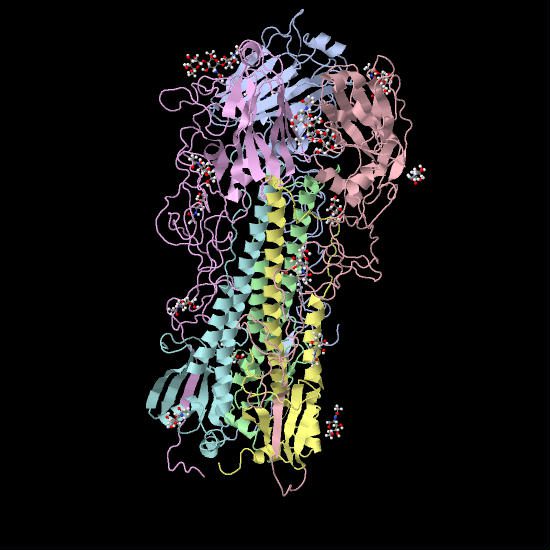
The sugar is actually quite tiny compared to the HA – it fits into a small pocket on the top of the spike. Here is a molecular model showing the HA bound to an analog of sialic acid. The globular top of the HA is at the top of the image. The tiny red and white spheres show where sialic acid would be bound, in a pocket at the top of the HA.

So far we have docked the influenza virion onto the surface of the cell. It’s sitting there quite firmly, but it’s still on the outside of the cell. How does it get in – or more accurately, how do the viral RNAs get into the cell? Stay tuned.

So what is the function of sialic acid (apart from allowing virus infection) – is it useful to us, or just an accident? Why is it there?
Sialic acids are in every cell and have many functions. A few
examples: Sialic acid-rich oligosaccharides on the glycoconjugates
found on surface membranes help keep water at the surface of cells.
Since water is a polar molecule with partial positive charges on both
hydrogen atoms, it is attracted to cell surfaces and membranes. This
also contributes to cellular fluid uptake. Sialic acid in the form of
polysialic acid is an unusual posttranslational modification that
occurs on the neural cell adhesion molecules, NCAM. In the synapse,
the strong negative charge of the polysialic acid prevents NCAM
cross-linking of cells.
Has there been any success to developing compounds that can out compete the HA ligand? Something that would bind stronger to sialic acid. I could see this as an attractive target for intervention.
Yes, some were developed years ago, but did not inhibit all HAs and
were never brought to market. However, sialic acid analogs have been
developed which inhibit the viral NA – these are the well known
antivirals oseltamivir and zanamivir. Sialic acid is the ligand for
the viral NA enzyme. I have an upcoming post on this topic.
Thanks very much for the reply, and for the whole blog – this is fascinating!
Pingback: Release of influenza viral RNAs into cells
Pingback: Souptopia
Pingback: Influenza HA cleavage is required for infectivity
If I understand correctly the Neuraminidase prunes sialic acid residues from glycoproteins. This is useful in allowing a clean get away after budding and also to stop binding of virions to each other. How does the virus prevent its NAs from removing the residues it needs to bind onto the glycocaylx before fusion? Wouldn’t its NAs be freeing it as fast as it could bind?
Always an interesting question. The idea is that during entry, the HA
binds and the particle enters before the slower-acting NA can remove
the sialic acid. This idea has some support from the HA assay;
initially virions bind red blood cells but after approximately 30
minutes the NA cleaves off sialic acid and reverses the HA. See
https://virology.ws/2009/05/27/influenza-hem….
Thanks but that then begs the question why when a new virion is budded does it not immediately bind and restart the fusion process before the NAs cut it free. On both occasions you have a virion adjacent to cell what is the difference why is there a net benefit to having NA cleaving sialic resisdues? There obviously is one or neuraminidase inhibitors would not work. I saw this http://www.ncbi.nlm.nih.gov/pubmed/19879239 and assumed it was due to nuraminidase having an inhibitory effect on cell infection although that effect was significantly outweighed by the benefits when budding
The NA protein is inserted into the plasma membrane before the virion
buds from the surface. Its presence may lead to removal of sialic
acids, which would prevent re-binding of the newly synthesized
particle. In theory at least; there are no data that directly answer
your question. The results in the paper you cite are consistent with
the idea that NA has some inhibitory effect during infection as would
be expected. Clearly there are significant differences between
infection and budding that are not fully understood.
Always an interesting question. The idea is that during entry, the HA
binds and the particle enters before the slower-acting NA can remove
the sialic acid. This idea has some support from the HA assay;
initially virions bind red blood cells but after approximately 30
minutes the NA cleaves off sialic acid and reverses the HA. See
https://virology.ws/2009/05/27/influenza-hem….
Thanks but that then begs the question why when a new virion is budded does it not immediately bind and restart the fusion process before the NAs cut it free. On both occasions you have a virion adjacent to cell what is the difference why is there a net benefit to having NA cleaving sialic resisdues? There obviously is one or neuraminidase inhibitors would not work. I saw this http://www.ncbi.nlm.nih.gov/pubmed/19879239 and assumed it was due to nuraminidase having an inhibitory effect on cell infection although that effect was significantly outweighed by the benefits when budding
The NA protein is inserted into the plasma membrane before the virion
buds from the surface. Its presence may lead to removal of sialic
acids, which would prevent re-binding of the newly synthesized
particle. In theory at least; there are no data that directly answer
your question. The results in the paper you cite are consistent with
the idea that NA has some inhibitory effect during infection as would
be expected. Clearly there are significant differences between
infection and budding that are not fully understood.
Pingback: Headless HA: Universal influenza vaccine?
The spheres in the picture should have opposite colors, the last sphere, the sialic acid should be orange, corresponding the color of the chair in the structures on the right.
is that right?
Thanks for picking that up – you are correct, the colors of the
spheres should be swapped. I have fixed the image.
Is it possible the newly formed HA bind to host proteins having sialic acid in Trans-Golgi network?
Possibly, although the presence of NA protein should remove sialic
acids from host proteins.
is sialic acid inhibitor of influenza virus
Â
there are reports that sialic acid is just a docking site not the receptor .why is it that we could not find  influenza virus receptor till date?
The sialic acids bound by influenza viruses are attached to proteins. The identity of the protein does not seem to matter for influenza virus entry. Hence sialic acid is the receptor.
Hello Prof. actually I love your blog very much. It helps me in understands better about the fascinating world of virology. I’m still an undergraduate student from Malaysia. Thanks prof. keep it up! ^^,
Prof, i like your illustration about attachment of the virion to cells. it help me to understand how influenza viruses bound to proteins
Are the receptors on the host cell always present and it just so happens that viral proteins can bind to them in a complimentary fashion? Viruses are parasitic so why do host cells have receptors for them to attach to?
This is very interesting. I was wondering do we have a clear idea about how  both HA and NA are already inserted into the plasma membrane before the budding takes place?Â
dear prof..i have read your entire blog…was so interesting… HA mediates viral binding and NA cleaves sialic acid linkage. i have one doubt regarding the resistance mechanism due to h274Y and N294s mutation, whats the reason behind the drug resistant mutation.
Pingback: The neuraminidase of influenza virus
Dear Prof, after reading this I came to have more questions about the process.
How long does a viron take to attach to a receptor of a cell. What happens if it fails to find a receptor? How long a viron survive if it fails to bind to any cell? Will a viron move on to the next target cell after trying to attach to the current cell for a while? Thanks.
Pingback: Virus Attachment Inhibitors - Remove Spyware, Malware and Viruses
Pingback: Crossing the species barrier – just one aminoacid to change? | StruBio
Dang!
Good questions!!!
You’re making me curious and you’re making me come up with a lot of “what ifs?!
Hi Prof. Racaniello, I have been watching the online course you posted on youtube. It very helpful! The glycoprotein underlying the sialyl glycans must be an endocytic protein, right? Otherwise the virus just stay attached outside. Could it be possible that some glycoprotein or glycolipids just serve as attachment factor that enrich the virus particle and there are some “true” receptors for the IAV?
And by “true” I mean the glycoproteins that are able to endocytose IAV.
Is it possible to make an “ideal layer” which can protect very single cell in our body from the recognition of the virus while the cells can still function normally, in the future? coz the virus keep mutating we can’t really overtake the mutation rate of them….but I know this idea is impossible in this century D: or even never ever. So good luck to everyone.
Which epithelial proteins does Influenza usually target?
hi i need to know the time taken by virus to bind to receptors before its entry to cell????