Understanding the transmission of SARS-CoV-2 is complicated by the large numbers of presymptomatic, asymptomatic, and mildly symptomatic (PAMS) patients. The reproductive number, R0, is a measure of population-level dynamics, but it cannot provide information on infectiousness of different groups such as PAMS subjects; when peak infectiousness occurs; and the effect of intrinsic properties of the virus. However information on infectiousness can be provided by a study of viral RNA load and infectivity in cell culture.
A study of 25,381 German COVID-19 cases was done to provide information on infectiousness during the course of infection. These included 6110 PAMS cases, 9159 hospitalized patients, and a series of times samples from hospitalized patients. Viral RNA loads – defined as RNA copies per swab – were determined by RT-PCR. The association between viral RNA load and probability of virus isolation in cell culture was estimated by using previously determined cell culture isolation data.
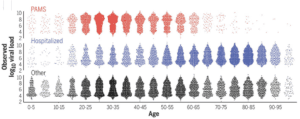
The results show that PAMS subjects have viral RNA loads (pictured) and predicted infectiousness, at the first positive test, slightly less than those of hospitalized patients. Children had similar viral RNA loads and predicted cell culture infectivity.
A small fraction of individuals’ first-positive viral RNA loads – 8%- had 10e9 RNA copies per swab. This observation is in line with previous findings that 15% of index cases are responsible for 80% of transmission, and that 8-9% of infected individuals carry 90% of the total viral load.
Time-course analysis revealed that viral RNA load peaks 1 to 3 days before onset of symptoms. From this peak, viral RNA loads decline 0.17 log10 units per day. Previous studies have shown that infectious virus is typically not recovered in cell culture beyond 10 days from symptom onset.
This study also examined viral RNA loads in 1533 patients with alpha variant infections. Compared with non-alpha infections, patients infected with the alpha variant had 10 times higher viral RNA loads and a 2.6 fold higher estimated cell culture infectivity. However, the impact of an increase in viral RNA load is, as the authors write, ‘dependent on context’ and the increase in cell culture infection probability is a ‘proxy indication of potentially higher transmissibility’. Complicating this analysis is the possibility that the correlation of viral RNA loads with cell culture infectivity might differ among variants.
The most important outcome of this study is that PAMS subjects – who in this study were tested at walk-in centers – are as infectious as hospitalized patients. As these individuals circulate in the community they can clearly trigger outbreaks of infection.

If 8% of patients had 10e9 copies of viral RNA per swab, that means that most COVID-19 patients must harbor at least 10e12 virus particles at some time during the course of their illness. That is a lot of virus. How does that number stack up against the prevalence of other highly infectious viruses such as measles or flu during the course of their illnesses? I know I saw this paper fly by several months ago but I have forgotten the authors. Would you provide a citation?
“ A small fraction of individuals’ first-positive viral RNA loads – 8%- had 10e9 RNA copies per swab.â€
This statement in paragraph 4 may be wrong. 10e9 means 10000000000, which is 10 times over the amount of RNA copies per swab detected in 8% of subject in the study.
Pingback: Estimate of infectiousness during COVID-19 - Virology Hub
10e9 is 10^9 a short billion or milliard
1 followed by 9 zeros, not 10 followed by 9 zeros.
It’s a shame phones don’t commonly allow superscript to show powers more clearly.
The original was indeed 10^9 .
However, 10e9 in most computer languages, in spreadsheets, or on old-time calculators does not mean 10 billion , it does in fact mean 10 x 10^9 = 10^10 as assumed by the commenter.
Unfortunately the medical community made a silly choice for unit notation years ago and it has persisted. Makes you wonder how many clinical trial spreadsheets out there are off by a factor of 10 …