
Bacteria use at least a couple of pathways for protecting themselves against viral infection. In return, multiple viral strategies for inhibiting or evading these pathways have evolved.
Two well-known bacterial strategies for self-defense against viruses involve immune enzymes, and roughly resemble the vertebrate innate and adaptive immune responses. Restriction enzymes, which recognize foreign DNA and selectively cut it up in precise locations known as restriction sites, are one example of such an immune enzyme. All pathogens harbor restriction sites in their genomes, and therefore this method serves as a non-specific or innate immune strategy because it does not depend on whether the pathogen was previously encountered. CRISPR-associated effector enzymes, for example Cas9, represent a second type of bacterial immune enzyme. CRISPR-mediated immune strategies rely on previous recognition and incorporation of viral genome segments into repeat clusters in the bacterial genome. These clusters are later transcribed and used to recognize and cleave newly invading viral genomes using specific effector enzymes. Because this method targets pathogens that were previously encountered, it is more similar to the vertebrate adaptive immune response.
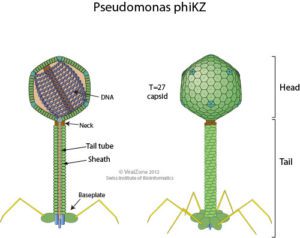
The bacterial species Pseudomonas aeruginosa is highly susceptible to infection by the giant bacteriophage ϕKZ (“phi kappa zeta”, illustrated, image credit), which in turn is highly resistant to P. aeruginosa’s immune enzymes. Interestingly, recent research shows that ϕKZ and phages like it assemble a nucleus-like shell after infecting a bacterial cell. This shell encloses the viral genome along with proteins involved in DNA replication, transcription, and recombination. The shell also separates these elements from the cytoplasm and proteins involved in translation. This separation of viral DNA from host cytoplasmic proteins led the authors of a recent Nature paper to hypothesize that the shell might shield viral DNA from bacterial immune enzymes and thus might be responsible for the broad resistance of ϕKZ to distinct bacterial immune responses.
To test this hypothesis, the authors infected P. aeruginosa cells with ϕKZ phages, and, using various fluorescence microscopy techniques, tracked the location of bacterial DNA-targeting CRISPR immune enzymes Cas9, Cas8, and Cas3, and bacterial restriction enzyme HsdR. Fluorescent images showed that each of these enzymes remained outside of the phage shell following phage infection, while phage DNA remained inside the shell.
To determine whether bacterial immune enzymes can cleave phage DNA if they can access it, the authors extracted ϕKZ phage DNA from virions and incubated it with Cas9 enzyme or with restriction enzymes EcoRI, HindIII, KpnI, or NcoI. Each of these enzymes cleaved the viral DNA under these in vitro circumstances, suggesting that naked ϕKZ phage DNA is susceptible to cleavage by immune enzymes.
As a next step in determining whether the shell protects phage DNA from bacterial immunity, the authors devised a strategy for smuggling various immune enzymes into the shell. First, they fused a Cas effector enzyme and a fluorescent enzyme to a phage-encoded protein that is able to traverse the shell membrane, ORF152. They transfected bacterial cells with this fusion construct, infected the cells with ϕKZ phages, and observed the combined effect of the Cas enzyme and viral infection by fluorescence microscopy. Surprisingly, the presence of the fusion construct had no effect on viral replication, and the fusion proteins were unable to traverse the shell membrane, likely because Cas enzymes are large.
The authors then chose EcoRI as an immune enzyme because it is much smaller than any of the Cas enzymes. They engineered three EcoRI fusion constructs:
- a wild type construct encoding ORF152 fused to EcoRI and a fluorescent enzyme;
- a mutant construct encoding the same ORF152/EcoRI/fluorescent enzyme fusion protein, but with a mutation in EcoRI that prevented it from cleaving DNA; and
- a negative control construct encoding EcoRI fused to a fluorescent protein, but lacking ORF152.
They transfected three different sets of P. aeruginosa cells with the respective constructs, infected them with ϕKZ phages, and analyzed them by fluorescent microscopy. As expected, the mutant construct localized inside the shell because it expressed ORF152, whereas the negative control was unable to cross the shell membrane because it lacked ORF152. In the P. aeruginosa cells transfected with ORF152 fused to wild type EcoRI, ϕKZ phages were prevented from forming full shells, and the phages did not complete the infection process. As a result, viral replication in these cells was markedly reduced compared to cells transfected with the other two constructs, i.e., mutant EcoRI constructs or constructs lacking ORF152. Cells harboring the wild type construct were also protected from phage-induced lysis, likely because viral replication was reduced. These results suggest that the shell protects the viral genome from bacterial immune enzymes.
The authors next hypothesized that although the viral DNA is resistant to bacterial DNA-targeting enzymes, the transcribed viral RNA might be sensitive to RNA-targeting enzymes once the RNA exits out of the shell and into the bacterial cytoplasm. As it turns out, Cas13a, such an RNA-specific CRISPR nuclease, was able to decrease ϕKZ phage replication by more than a million fold, suggesting that ϕKZ is susceptible to RNA-specific immune enzymes.
The assembly, during virus infection, of a shell that roughly resembles a nucleus raises several questions. Does the shell purely serve as a mechanism of evading host cell immune defenses? Or, is it a remnant of a cellular structure, thereby suggesting that the virus may originate from an ancient parasitic cell? Or, was assembly of a shell a move towards further independence from host cells? It is hard to answer any of these questions, but at the very least, this study provides more insight into the continuous evolutionary dance between viruses and bacteria.
