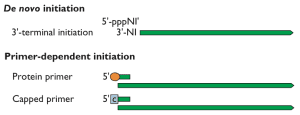

The requirement for a primer in the initiation step of nucleic acid synthesis varies among the different classes of polymerases. All DNA polymerases are primer-dependent enzymes, while DNA-dependent RNA polymerases initiate RNA synthesis de novo – without a primer. Some RNA-dependent RNA polymerases can also initiate RNA synthesis without a primer: the enzyme begins by adding the first base complementary to the template RNA (illustrated). Other RNA-dependent RNA polymerases require a primer to initiate synthesis. Examples shown on the illustration include the protein-linked primer of picornaviruses, which consists of the protein VPg covalently attached to two U residues. The primer for influenza virus mRNA synthesis is a capped oligonucleotide 12-14 bases in length that is cleaved from the 5€² end of cellular mRNA.
The structure of the RNA-dependent RNA polymerase of hepatitis C virus reveals how a primer-independent RNA polymerase positions the first nucleotide on the RNA template. This process is illustrated below. With the RNA template (dark green) in the active site of the enzyme (panel A), a short beta-loop (brown) provides a platform on which the first complementary nucleotide (light green) is added to the template. The second nucleotide is then added (panel B), producing a dinucleotide primer for RNA synthesis. At this point nothing further can happen because the priming platform blocks the exit of the RNA product from the enzyme (panel B). The solution to this problem is that the polymerase undergoes a conformational change that moves the priming platform out of the way and allows the newly synthesized complementary RNA (panel C, light green) to exit as the enzyme moves along the template strand.
The structure of the RNA polymerase of hepatitis C virus reveals that it is not really a primer-independent enzyme: a dinucleotide primer is synthesized by the polymerase using a protein platform in the active site. Such protein platforms also appear to be involved in the priming of RNA synthesis by other flaviviruses (dengue and West Nile viruses), influenza virus (genome RNA synthesis is primer independent), reovirus, and bacteriophage phi6. Perhaps all viral RNA-dependent RNA polymerases are dependent on such priming platforms to initiate RNA synthesis.

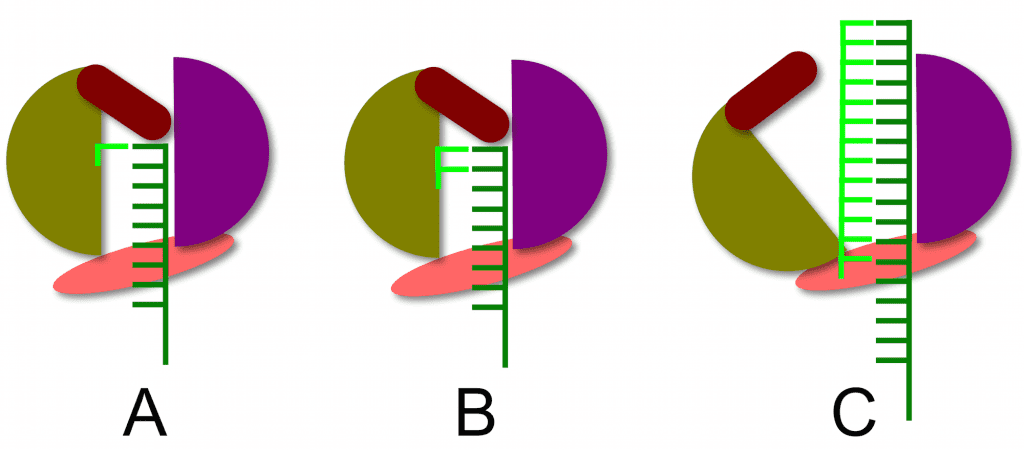
From the structures it also looks like these platforms (both Hep C and Phi6) have tyrsoines which point towards the active site of the polymerase. In these cases they stack with the first base of the nascent chain but the OH group of the tyrosine is very close to the active site. It begs the question is has VPg based protein priming evolved from this system (or vise versa)?
PS I don’t know enough about the topic to know if I am stating the obvious here.
I see what you mean (Fig. 1B, C in the paper). Your idea is reasonable, although I don’t know how a part of the pol would become a protein primer. Good idea though.
Pingback: TWiV 330: A swinging gate