
Sequence Searcher is a Java program that allows users to search for specific sequence motifs in protein or DNA sequences. For example, it can be used to identify restriction enzyme cleavage sites or find similar sequence patterns among multiple sequences. Most searches run in a few seconds.
Sequence Searcher is part of the Virology.ca suite of programs available at the University of Victoria.
Help files:
Some of the key features of Sequence Searcher include:
- Searching through multiple sequences
- Use of regular expressions or fuzzy search patterns.
- Searching for patterns on both strands of a DNA sequence
- Graphical representation of results and ability to save search results
- It can run on multiple computer platforms (Java)
For DNA, the searches are conducted by finding the motif within a sequence from the 5€™ to 3€™ end on the top strand. The searches are also processed from the 5€™ to 3€™ end of the bottom strand. As a result, bases are numbered from 1 starting at the 5€™ at either the top or bottom strand.
Regular expression and fuzzy pattern searches are available:
Fuzzy searches provide the option for the program to allow a certain number of mismatches from a sequence input at any position. Note that the maximum number of mismatches that the program allows is 40% of the length of the sequence motif.
Regular expression allows for inputs of precise motifs along with considerable user-specified flexibility at specific positions.
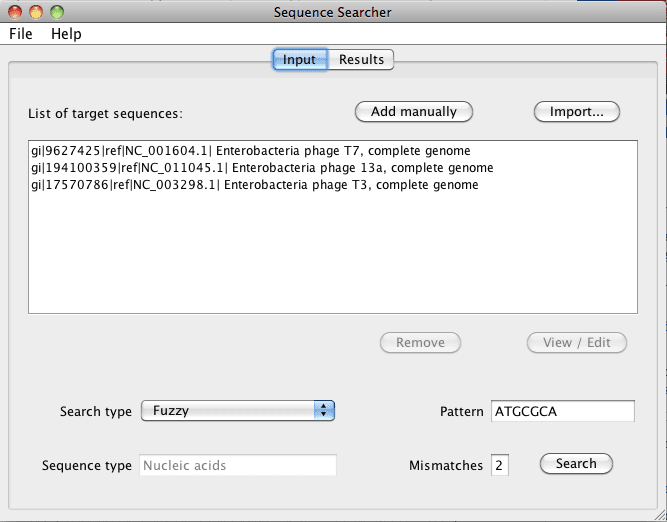
Figure 1. The input tab is where you can import DNA or protein sequences (must be in FASTA format) and type in the specific pattern to search within in the sequence(s). The search type can be selected as Regular expression€ or Fuzzy€ by using the drop down menu.
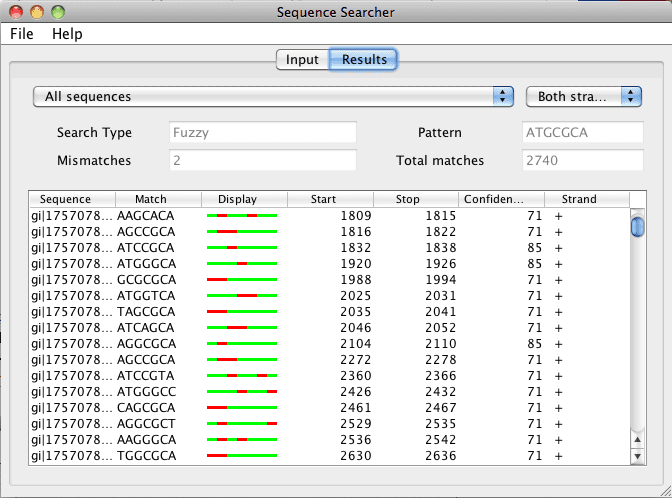
Figure 2. When a search has been completed, the results tab is presented in a table format. The results in the table can be sorted depending on the column header (sequence, match, start, stop, confidence, and strand). The results can also be filtered by sequence and strand by selecting the drop down menus at the top.
Marass, F., & Upton, C. (2009). Sequence Searcher: A Java tool to perform regular expression and fuzzy searches of multiple DNA and protein sequences BMC Research Notes, 2 (1) DOI: 10.1186/1756-0500-2-14