
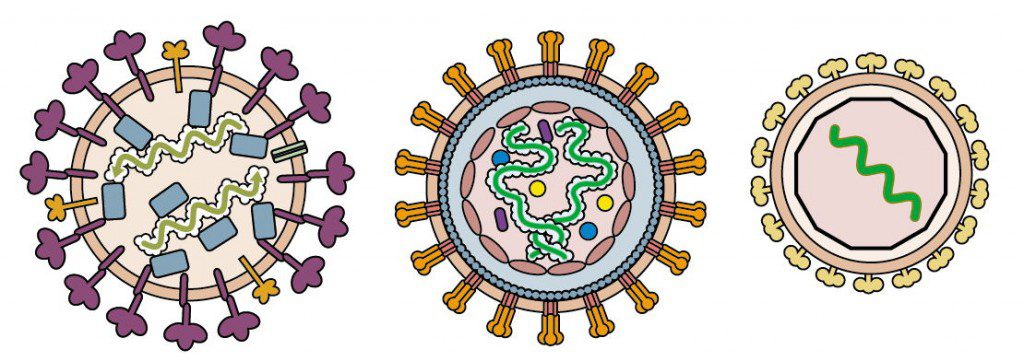
Examples of enveloped virions that contain nucleoproteins are shown in the figure below. These include influenza virus (left), a simple retrovirus (center), and a togavirus (right).
The influenza virion contains segments of viral RNA bound to four different proteins. Retroviral RNA is bound to a nucleocapsid protein which in turn is enclosed in a capsid, while togavirus RNA is located within an icosahedral shell.
Until recently, it was believed that the genome of all other known enveloped DNA and RNA viruses is always associated with one or more viral proteins. This belief may be changed by the isolation, from a solar saltern in Trapani, Italy, of a virus that infects the archaeal species Halorubrum. Salterns are multi-pond systems in which sea water is evaporated to produce salt. In such hypersaline envrionments, Archaea predominate, and about 20 archaeal viruses have been isolated from these locations.
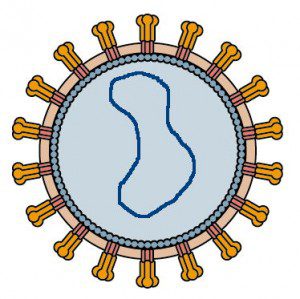
The virus isolated from the Italian saltern is called Halorubrum pleomorphic virus-1, or HRPV-1. Biochemical analyses of the virion show that it is composed of lipids and two structural proteins, VP3 and VP5. The genome is a circular ssDNA about 7 kb in length with nine open reading frames. The virion architecture is unique: it is composed of a flexible membrane (hence the designation pleomorphic) that contains external spikes of the VP4 protein, and is lined on the interior with VP3. The viral DNA is apparently not bound to any proteins in the virions.
At the upper left is my depiction of the appearance of HRPV-1. The diagram was produced by deleting the internal proteins and nucleic acid of a simple retrovirus and replacing these with a ssDNA genome. The HRPV-1 VP4 spikes and the internal VP3 proteins are present, but no proteins are bound to the viral genome. Whether or not the VP4 spikes are oligomeric as shown is unknown.
Most enveloped viruses acquire their lipid membrane by budding from the host cell, and a similar mechanism could account for the formation of HRPV-1 virions. In the absence of a nucleoprotein, it is not clear how the viral genome would be specifically incorporated into the budding envelope. Another condundrum is how the virions would pass through the proteinaceous layer that covers the archaeal host cell.
Whether HRPV-1 is representative of a new kind of virus lacking nucleocapsid protein will be revealed by the study of other pleomorphic enveloped viruses. Candidates include bacterial viruses that infect mycoplasmas, and another pleomorphic haloarchaeal virus isolated from a different Italian saltern, Haloarcula hispanica pleomorphic virus 1.
Pietila, M., Laurinavicius, S., Sund, J., Roine, E., & Bamford, D. (2009). The Single-Stranded DNA Genome of Novel Archaeal Virus Halorubrum Pleomorphic Virus 1 Is Enclosed in the Envelope Decorated with Glycoprotein Spikes Journal of Virology, 84 (2), 788-798 DOI: 10.1128/JVI.01347-09

Amazing. Do the authors of the paper propose any hypotheses about how the virus manages to get out and into the cell?
Just my pedantry: “a bacteriophage that infects the archaeal species Halorubrum”. Hmm… “bacteriopage” doesn't sound right to me. A few people have used the terms “arch(a)eophages” or “arch(a)eoviruses” in the past but “archaeal viruses” seems the safest choice, I think.
(Actually, I was thinking now that the common terms “bacteriophage” and “phage” are a little misleading — some people may actually think that the “real viruses” are only those of eukaryotes!)
You are absolutely correct; bacteriophage is the wrong term to use to describe the Halorubrum virus; it is a virus. That was entirely my error, and surely reflects the number of years I have been in the field! I agree that we should call all viruses by that name only; bacteriophage of course is an old term and as such remains ensconced in the literature. A virus is a virus; they are all real.
As for how the Halorubrum virus enters and exits cells: the authors have little to say on either subject. One would expect that on entry the viral membrane would fuse with that of the host cell, but how that happens is unknown given the proteinaceous coat of the archaeal host cell. Exit by budding presents a similar problem. The authors admit that they have never seen such viruses 'budding' from host cells.
It is interesting that the archaeal host cells are not adversely affected by infection with the virus; they continue to divide normally and do not lyse. Nevertheless, it is possible to titrate the virus on lawns of Halorubrum by plaque assay.