The plaque assay is a terrific method for determining virus titers, but it doesn’t work for all viruses. Fortunately there are several alternative methods available, including the end-point dilution assay.
The end-point dilution assay was used to measure virus titer before the development of the plaque assay, and is still used for viruses that do not form plaques. Serial dilutions of a virus stock are prepared and inoculated onto replicate cell cultures, often in multi-well formats (e.g. 96 well plastic plates). The number of cell cultures that are infected is then determined for each virus dilution, usually by looking for cytopathic effect.
In this example of an end-point dilution assay, 10 monolayer cell cultures were infected with each virus dilution. After an incubation period, plates that displayed cytopathic effects were scored with a +. At high dilutions, none of the cell cultures are infected because no particles are present. At low dilutions, every cell culture is infected. Half of the cell cultures showed cytopathic effects at the 10-5 dilution. This is the end point: the dilution of virus at which 50% of the cell cultures are infected. This number can be calculated from the data and expressed as 50% infectious dose (ID50) per milliliter. The virus stock in this example contains 105 ID50 per ml.
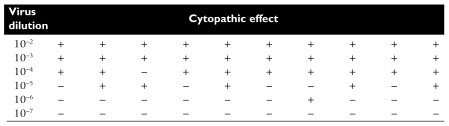

In real life, the 50% end point does not usually fall exactly on a dilution as shown in the example. Therefore statistical procedures are used to calculate the end point of the titration.
End-point dilution methods can also be used to determine the virulence of a virus in animals. The same approach is used: serial dilutions of viruses are made and inoculated into multiple test animals. Infection of the animal can be determined by death or clinical symptoms such as fever, weight loss, or paralysis. The results are expressed as 50% lethal dose (LD50) per ml or 50% paralytic dose (PD50) per ml when lethality or paralysis are used as end points.
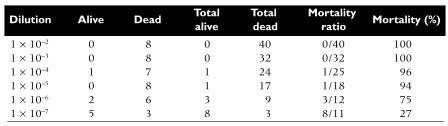
The following example illustrates the use of end point dilution to measure the lethality of poliovirus in mice. Eight mice were inoculated per virus dilution, and the end point was death. The statistical method of Reed and Muench was used to determine the 50% end point. In this method, the results are pooled, and the mortality at each dilution is calculated. The 50% end point, which falls between the fifth and sixth dilutions, is calculated to be 10-6.5. Therefore the virus sample contains 106.5 LD50 units.

Reed, L.J., & Muench, H. (1938). A simple method of estimating fifty percent endpoints. Am. J. Hygiene, 27, 493-497

The statistical method of Reed and Muench is really a complicated one and one should have good mathematical skills to calculate TCID50. I developed my own formula (even layman can calculate TCID50 by my formula and it is 100% match with Reed and Muench or modified Karber formula). I am willing to share my formula if anybody interested. Please contact me by my email.
Thanks, rama@umn.edu
Well, it seems that the developed formula by Dr Rama, can work good! however I've not use it yet!
As soon as test it I'll tel the result
Good luck
Dear Sir,
I thank you for posting this informative note in the website. May I request you to present ur views on my problem which i face while doing LD50 calculation using reed and muench method. I want to have number of organisms as LD50 units. The usual method given by Dr. reed gives the value of exact dilution of the seed which will be equivalent to 1 LD50 unit. Pls educate me how its possible to have these units in terms of number of microbes (pfus or CFUs).
Thanx and reggards
mayank
Dear Sir,
I thank you for posting this informative note in the website. May I request you to present ur views on my problem which i face while doing LD50 calculation using reed and muench method. I want to have number of organisms as LD50 units. The usual method given by Dr. reed gives the value of exact dilution of the seed which will be equivalent to 1 LD50 unit. Pls educate me how its possible to have these units in terms of number of microbes (pfus or CFUs).
Thanx and reggards
mayank
Dear Sir,
I thank you for posting this informative note in the website. May I request you to present ur views on my problem which i face while doing LD50 calculation using reed and muench method. I want to have number of organisms as LD50 units. The usual method given by Dr. reed gives the value of exact dilution of the seed which will be equivalent to 1 LD50 unit. Pls educate me how its possible to have these units in terms of number of microbes (pfus or CFUs).
Thanx and reggards
mayank
Mayank! Please send me your email address . My ID is rama@umn.edu
Dr. Rama's formula is easy to understand and super easy to use. Thanks
It is straightforward to calculate the LD50 as the concentration of
the stock virus times the 50% end point titer. For example, if we have
a stock of virus with a titer of 1x10e9 pfu/ml, and the LD50 is
calculated to be 10e-6.5, then by multiplying the two numbers we
calculate that the LD50 is 3x10e2 PFU. The same may be done for CFUs.
do you know where I can get a free digital copy of Reed, L.J.; Muench, H. (1938). “A simple method of estimating fifty percent endpoints”. The American Journal of Hygiene 27: 493–497?
Thanks,
JON
Thanks for the formula–much appreciated.
Nick
Jon, I've looked all over and can't find a copy, not even in my files.
And our library just removed all its copies of old journals. Sorry.
How about determining the LT50?
Do you have formula in solving thr LT50?
Actually, I have never understood the utility, or even the validity, of pooling the results at each dilution.
In the example shown above, why is the mortality ratio of 27% at 1×10-7 , calculated from the cumulative data considered to be more valid than the actual observed mortality ratio at that dilution (3/8 or 37.5%) ?
I have read (and even tried to understand) textbook explanations of this, and I have not found the explanations very clear.
DMc
Have you read the 19389 paper by Reed and Muench? The logic is best
explained there. If you have not, I can send you a copy of the paper.
Do you mean LD50? Then Yes I have
No, I haven't. I think about the only paper I have ever read from that period was the Ellis and Delbruck paper on the single-step growth curve. (Even with that one, though, the credit goes to Alan Cann for putting it on the CD that came with his text book). If you could send me a copy, I would be much obliged. I'll even write back here to let you know if I understood it better this time.
DMc
OK, after reading the classic paper, I think I have a (slightly) better understanding of why using cumulative numbers of dead mice/infected wells and cumulative alive mice/non-infected wells works. In the actual paper, the example they gave was protection of mice from infection by transfer of different dilutions of immune serum. However, to make this relevant to viral titres in cell culture, I will try to explain how I understood the paper as if the data were referring to a TCID50 determination .
I found the figure much clearer than the text in explaining the use of cumulative numbers. If you plot the cumulative number of infected wells going from the most dilute to the least dilute, then you get a curve that goes down as the inoculum gets more dilute. Conversely, the curve for cumulative number of non-infected wells goes up as the inoculum gets more dilute. Where the two curves cross is the dilution that gives you the TCID50.
However, they didn't use these two curves to calculate the TCID50. This was done using the Reed+Muench formula on the sigmoid curve of cumulative or pooled % of infected wells at each dilution. I guess that finding where you get 50% infected wells on this curve is identical to finding where the number of infected/non-infected wells is the same.
OK, so now I understand why it's a valid approach. But what is the utility? Well if I understood correctly, using cumulative numbers is supposed to be more statistically robust, since the number of animals/cell culture wells analyzed at each dilution is quite low (typically 6 to 8). If this is correct, then the variance of a TCID50 measure using the Reed-Muench method correctly (i.e. with cumulative numbers) should be lower than if the Reed-Muench formula is applied directly to the observed proportion of infected wells at each dilution.
It occurs to me that I could test this quite soon, as I will be getting a bunch of write-ups from students who did a TCID50 in the lab this semester, so I have a data set with several determinations on the same virus stock.
DMc
Dear profvrr,
can you send me a copy, i am interested in Reed an Muench method,because it is too early ,i can not search it in our library. thank you !
Dear Dr Vincent,
My study virus disappears in the process of passaging (after 3 passages) and is replaced by another unknown CPE producing virus on LLC-MK2. I cannot understand that how do I identify this unknown virus and select my original virus. Since i need to prepare the viral stock for my original virus. The approval of cloning based detection methods would take around 2-3 months. In passage 2 original virus is there and in the passge 3 it disappears. Perhaps the exclusion principal is involved here. SHould i try even more cell lines or the available antibodies. Any thoughts or golden piece of advice? Yoroshiku one gai shimas.
Dear Dr Vincent,
My study virus disappears in the process of passaging (after 3 passages) and is replaced by another unknown CPE producing virus on LLC-MK2. I cannot understand that how do I identify this unknown virus and select my original virus. Since i need to prepare the viral stock for my original virus. The approval of cloning based detection methods would take around 2-3 months. In passage 2 original virus is there and in the passge 3 it disappears. Perhaps the exclusion principal is involved here. SHould i try even more cell lines or the available antibodies. Any thoughts or golden piece of advice? Yoroshiku one gai shimas.
Dear Vincent,
I am a master student in Biotechnology from Taiwan. I am looking for the paper by Reed and Muench on the internet and our school library, however, I can not find it. May I have the paper from you? Appreciate for your help.
DMc,
IT'S OBVIOUS FROM YOUR COMMENT THAT YOU'VE DIGESTED THE CLASSIC OF REED AND MUENCH. PLEASE, CAN YOU SEND ME A COPY?
faleyetope@yahoo.com
ps, I'm a graduate student of virology in Nigeria. The logic behind the calculation still eludes me. Thanks.
I would try either a different cell line, hoping that it will only
propagate your virus, or try diluting the virus before infecting
LLC-MK2 so that only a few virions are used to infect, thereby
favoring infection with one or the other virus. If your virus is in
the majority this should work, especially if done a few times.
Thanks Dr Vinc, Your advise was quite useful in enabling me to select samples containing only single virus. Recently I tried to determine the replication kinetics of many different strains of the same virus. I found out that the replication of the viruses was quite low and eventually became undetecable by qPCR after 04 passages on LLC-MK2 with inapparent CPE. What would you advise in case that I try antagonizing the IFN, can it lead to any improvement in the replication?
Yes, it's a good idea to try to disrupt IFN production. You could try
another cell line with an IFN defect (such as VERO, which don't
produce IFN, or cells lacking the type I IFN receptor), or you could
alter the LLC-MK2 so they are defective in the IFN response.
I am interested to have a read of this paper. May I have a copy from you? Appreciate.
PLS SEND ME UR CALCULATION FORMULA
Dear Vincent,
I am a master student in Biotechnology from Vietnam. I am looking for the paper by Reed and Muench on the internet and our school library, however, I can not find it. May I have the paper from you? Appreciate for your help.
Hi Dr. Racaniello,
I am also looking for the Reed and Muench, I would very much appreciate if I can obtain the paper from you. Thanks a lot.
cyc
Respected sir,
I would like to know weather TCID50 can be used to evaluate antiviral activity of compounds.
regards
Prashant,PhD
Hi Vincent,
I have a technical question about the TCID50 method. I have to assay lots of samples from infected mice and I set them up in quadruplicate so I usually have a lot of 96well plates to work with(20-30 at a time). After absorbing virus on MDCKs for 24 hrs I replace the media with VGM by aspiration. Aspirating the media off is very time consuming and uses up a lot of tips. I was wondering if I could save time by “flicking” the media off of my plates into a container instead of aspirating. Any tips for saving time would be helpful.
Yes, you can flick the medium off of the 96 well plates – use a broad
tray to receive the liquid. You can flick the plate with your wrist,
or tap it – try it on some blank plates to get a feel for what works
for you. We do this all the time when changing medium in 96 well
plates – as you say, it takes too long to remove the medium by
aspiration.
Dear Dr,
I’m a PhD student and I would be interested in having a copy of this paper, could you send it to me please?
Thanks a lot.
Hello,
Could you please send me your formula?
Best regards
Pingback: XMRV infection of Rhesus macaques
please send me your formulae
Elisha kutto
yonkutt@yahoo.com
your comment seems can solve my problem which has been an headage for me. please clarify for me. what is the difference between the 50% end point titre and the ld50.how can i calculate the number of organisms (cfu) per ld50?. must i know the concentration of the stock solution to determine the ld50?
Thank you,elisha yonkutt%yahoo.com
Hello
please send me you formulae
I am an MSc student of clinical pathology from Kenya.I would be very gratefull if you send me the paper by Reed and Muench.Thank you
Dear profvrr
i will apreciate if you send me a copy of Reed and Muench paper
elisha kutto
yonkutt@yahoo.com
please send your email address to maramakrishnan@gmail.com
Please send your email address to maramakrishnan@gmail.com
Hi,
I’m a PhD student struggling to understand the whole purpose of the TCID50. I have calculated using the Reed and Muench method that my virus’ TCID50 is 5.1E-7. In the experiment I did a series of 10 fold dilutions, set up by adding 0.3ml virus to 2.7ml medium etc etc. I then put 100microlitres of each dilution into wells containing 100microlitres of cells in medium. I did twelve wells per dilution. So how do I know what my TCID50/ml is? Or is the value I calculated already per ml? And what units is it expressed in i.e what does it actually mean?! I have previously calculated the titre of the virus using plaque assay, how do the two relate? I am succeeding in getting myself more and more confused the more I read, so any help much appreciated! Thanks, KC
Not sure what you are asking – please email virology@virology.ws and we’ll figure it out.
Hi,
I work in an animal disease lab, where we use TCID50 to determine virus use in VN/SN testing. For the past few months there has been some differing opinions on the best way to measure the TCID50 units. After receiving information from Dr. Rama, everyone involved is in agreement that his formula is the most accurate and less complicated way to measure.  I am very grateful to Dr. Rama for emailing his formula to me. Thanks Again, Dr. Rama
M DeniseÂ
hello rama please send your formula, my mail id is senvetpath@gmail.com
 Vincent sir,Â
I am Ph.D student working on virus. Kindly end me the Reed and Muench original paper on end point dilution assay for virus titer. My email id is sonal.mundhra@gmail.com
Thank you.Â
dear sir can you please mail me the same artical i dont have online access.
dav0082@gmail.com
Thanks a lot sir for the paper.