Now that we have examined influenza viral RNA synthesis, it’s a good time to step back and look at a very important property of this step in viral replication. All nucleic acid polymerases insert incorrect nucleotides during chain elongation. This misincorporation is one of the major sources of diversity that allows viral evolution to take place at an unprecedented scale. Put another way, viruses are so successful because they make a lot of mistakes.
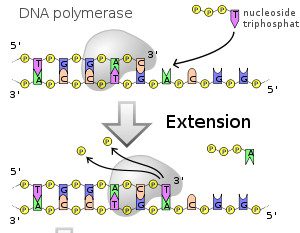
Nucleic acids are amazing molecules not only because they can encode proteins, but because they can be copied or replicated. Copying is done by nucleic acid polymerases that ‘read’ a strand of DNA or RNA and synthesize the complementary strand. Let’s start by examining DNA synthesis. Below is a DNA chain, which consists of the bases A, G, C or T strung together in a way that codes for a specific protein. In this example, the template strand is at the bottom, and consists of the bases A, C, C, T, G, A, C, G, and G (from left to right). A DNA polymerase is copying this template strand to form a complementary strand. So far the complementary bases T, G, G, A, and C have been added to the growing DNA chain. The next step is the addition of a T, which is the complementary base for the A on the template strand:
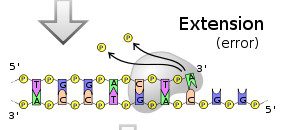
So far all is well. But all nucleic acid polymerases are imperfect – they make mistakes now and then. This means that they insert the wrong base. In the next step below, the DNA polymerase has inserted an A instead of the correct G:
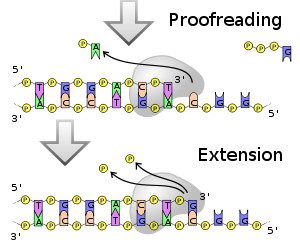
Insertion of the wrong base leads to a mutation – a change in the sequence of the DNA. In general, it’s not a good idea to make new DNAs with a lot of mutations, because the encoded protein won’t function well (but there are exceptions, as well will see). But in this case, there is a solution – DNA-dependent DNA polymerases (enzymes that copy DNA templates into DNA) have proofreading abilities. The proofreader is an enzyme called exonuclease, which recognizes the mismatched A-C base pair, and removes the offending A. DNA polymerase then tries again, and this time inserts the correct G:
Even though DNA polymerases have proofreading abilities, they still make mistakes – on the order of about one misincorporation per 107 to 109 nucleotides polymerized. But the RNA polymerases of RNA viruses are the kings of errors – these enzymes screw up as often as one time for every 1,000 – 100,000 nucleotides polymerized. This high rate of mutation comes from the lack of proofreading ability in RNA polymerases. These enzymes make mistakes, but they can’t correct them. Therefore the mutations remain in the newly synthesized RNA.
Given a typical RNA viral genome of 10,000 bases, a mutation frequency of 1 in 10,000 corresponds to an average of 1 mutation in every replicated genome. If a single cell infected with poliovirus produces 10,000 new virus particles, this error rate means that in theory, about 10,000 new viral mutants have been produced. This enormous mutation rate explains why RNA viruses evolve so readily. For example, it is the driving force behind influenza viral antigenic drift.
Here is a stunning example of the consequences of RNA polymerase error rates. Tens of millions of humans are infected with HIV-1, and every infected person produces billions of viral genomes per day, each with one mutation. Over 1016 genomes are produced daily on the entire planet. As a consequence, thousands of mutants arise by chance every day that are resistant to every combination of antiviral compounds in use or in development.
I cannot overemphasize the importance of error-prone nucleic acid synthesis in RNA viral evolution and disease production. We’ll spend the next two days examining the consequences of error prone replication. First we’ll consider the implications for viruses as a population, and then we’ll discuss the outcome when a virus produces an RNA polymerase that makes fewer mistakes. Please bear with me as we diverge slightly from influenza virus; these concepts will be an important and enduring component of your toolbox of virology knowledge.

Mind blowing. Again, excellent reading!
Pingback: Ascription is an Anathema to any Enthusiasm › Searching the Alternate Routes
This isn't my field (I'm a software guy), but I have a certain fascination with your field. I say this, because I have a speculation I'd like to throw out there, but it's likely kinda ill informed. Nevertheless, I'm curious as to what you might think.
You refer to this viral RNA synthesis as error-prone, which I understand what you mean when you say this. But the word “error” carries with it a certain deviation from “correctness”, at least in common parlance. In the viral RNA synthesis case, might this “error rate” actually be a primary survival mechanism for the virus in the sense that it helps its genome evade immune system defenses and anti-viral molecules.
If so, might there be a drug strategy which instead of targeting the replication cycle of the virus, instead attacks its error rate. The idea being that by lowering the error rate of the viral RNA synthesis one reduces the rate at which the virus produces immune-evading, or anti-viral drug evading mutations. Supposing one found this “exonuclease as a drug” molecule, you could administer it seasonally to make this year's vaccine more likely to be effective on successive years, or something like that.
But I am an amateur, so I'm probably full of crap.
You are not at all full of crap; in fact those are excellent ideas.
Often amateurs (or should we say, those not in the field) have
excellent ideas because they are not saddled with the baggage of
familiarity.
As you'll see in coming posts, in fact the 'errors' do help the virus
to survive, by allowing it to adapt to new hosts, to evade antivirals,
immunue responses, etc. But only to a certain extent – if there are
too many errors they will interfere with viral replication.
As for your idea of lowering the error rate as an antiviral approach –
you'll see in the coming posts that this can be done experimentally,
and it has dramatic consequences on viral fitness. On the other hand,
what about pushing the error rate the other way, so that you
mutagenize the virus out of existence? This is in fact the mode of
action of at least one antiviral drug, ribavirin, which we'll discuss
here in a few days.
Good article, great illustrations, once again.
Apparently, RNA viruses form quasi-species because they have a mutation rate similar to the 1/(number of bases)
in their genome. In the case of DNA organisms, the DNA polymerase reduces this mutation rate to 1/10^7 – 1/10^9.
This is also the order of magnitude of their genome. This would mean that many DNA organisms (including humans) would form quasi-species if all its genome were codifying. As a consequence, a possible role for
introns would be to allow for unharmful mutations in order to stabilize the species. Is this argument right?
thanks Vincent for the blog – I've enjoyed following the discussions
Here's a tough question. In the follow up blog to this, you say that the high mutation rates of RNA viruses is beneficial to survival in a complex environment. If this is true, why don't DNA viruses evolve high mutation rates also? It would be simple for them to delete their proofreading domain
This is a fabulous question. I'll speculate because there is no known
answer, as far as I know. The known RNA viral genomes are no longer
than 27-31 kb, probably because they would sustain too many lethal
mutations if they were longer. DNA viral genomes can be up to 1.2
million bases in length – probably because they have error correction
mechanisms. So I would speculate that DNA viruses have evolved to keep
error repair pathways so that the genomes can be longer. Smaller DNA
viruses don't have their own DNA polymerases, but use those of the
host. Host DNA systems need to have error repair, otherwise they will
sustain mutations that lead to diseases such as cancer. Both forms of
reproduction are evolutionarily sustainable: shorter RNAs with lots of
errors; longer DNAs with fewer errors.
I don't want to go to far afield here, but when did error repair first begin? Are there more primitive forms of error repair to be contrasted with extremely sophisticated ones? Is that a distinguishing difference between RNA and DNA viruses, i.e. the former always lacks error repair mechanisms but the latter generally contains them?
Interesting stuff. Shannon and Weaver's Mathematical Theory of Communication 1948, which is the seminal work that founded electrical engineering (and indeed made the modem possible) is all about efficiently reducing error rates in a noisy channel (stop bits and parity bits).
Here is a link to that 1948 paper which led to the birth of information theory and electrical engineering.
http://cm.bell-labs.com/cm/ms/what/shannonday/s…
Notice on page 6 how similar their coding schema is to CG AT CG GC AT, etc!
An interesting question. According to wikipedia: The fossil record
indicates that single celled life began to proliferate on the planet
at some point during the Precambrian period, although exactly when
recognizably modern life first emerged is unclear. Nucleic acids
became the sole and universal means of encoding genetic information,
requiring DNA repair mechanisms that in their basic form have been
inherited by all extant life forms from their common ancestor. The
emergence of Earth's oxygen-rich atmosphere (known as the “oxygen
catastrophe”) due to photosynthetic organisms, as well as the presence
of potentially damaging free radicals in the cell due to oxidative
phosphorylation, necessitated the evolution of DNA repair mechanisms
that act specifically to counter the types of damage induced by
oxidative stress.
In general we believe that RNA viruses lack error correction
mechanisms, but there may be some exceptions. For example, there is
some evidence that a coronavirus might have error correction
machinery.
An interesting question. According to wikipedia: The fossil record
indicates that single celled life began to proliferate on the planet
at some point during the Precambrian period, although exactly when
recognizably modern life first emerged is unclear. Nucleic acids
became the sole and universal means of encoding genetic information,
requiring DNA repair mechanisms that in their basic form have been
inherited by all extant life forms from their common ancestor. The
emergence of Earth's oxygen-rich atmosphere (known as the “oxygen
catastrophe”) due to photosynthetic organisms, as well as the presence
of potentially damaging free radicals in the cell due to oxidative
phosphorylation, necessitated the evolution of DNA repair mechanisms
that act specifically to counter the types of damage induced by
oxidative stress.
In general we believe that RNA viruses lack error correction
mechanisms, but there may be some exceptions. For example, there is
some evidence that a coronavirus might have error correction
machinery.
Pingback: Pandemic H1N1 influenza virus outcompetes seasonal strains in ferrets
Pingback: This Week In Virology – Esta Semana en VirologÃa « Gripe por A (H1N1) Blog
Wow…this was a very nice thing to my article discussion on CTV (Citrus tristeza virus)…thanks a lot
I'm studying medicine and your diagrams were very helpful.
great! very useful information.
where do you get all those diagrams from? Do they exist in vectorial format as well?
Simple and clear, thank you.
This is a good article but today I read something new, I read that RNA pol do have proofreading activity (http://mbe.oxfordjournals.org/content/22/6/1444.full.pdf).
Article says that RNA pol have proofreading activity but when I saw the references they gave I was not fully convinced as there are transcription factors etc which are facilitating these proof readings.
So can someone help me to clear up my mind??
Pingback: What we are not afraid to say about Ebola virus
Pingback: Ebola spreading? - Page 2 - Christian Chat Rooms & Forums
Pingback: Lessons from the Flu
If the sequence of RNA changes then definitely the resultant protein will b changed,. HOW then it is possible that despite of such mutation same kind and type of proteins build
Pingback: Od polio chcę mieć Ukrainę wolną | Sporothrix
Pingback: A huge host contribution to virus mutation rates