Once the (-) strand influenza viral RNAs enter the nucleus, they serve as templates for the synthesis of mRNAs. These molecules are then transported back to the cytoplasm, where they direct the synthesis of viral proteins. However, the mRNAs are not complete copies of the viral (-) strand RNAs – they are missing sequences from both the 5′- and 3′-ends. Therefore, to produce more viral (-) strand RNAs that are needed to assemble new virions, a full length (+) strand is produced, which in turn is copied to a full-length (-) strand RNA. The (-) strand RNAs are then used to assemble new virions. This process is best understood by referring to the figure, which distinguishes the processes of mRNA synthesis and replication. The (-) strand RNAs are those which are present in virions. The same processes occur for all eight segments of influenza viral RNA.
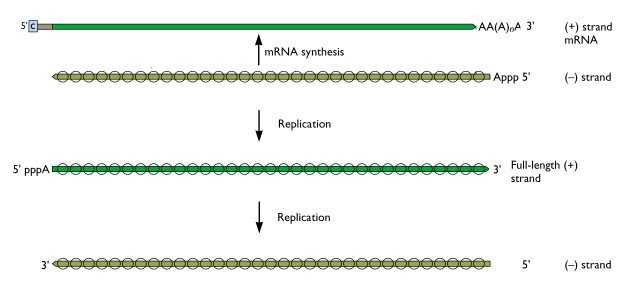

The enzyme that reproduces influenza RNA is known as an RNA-dependent RNA polymerase. This enzyme, which consists of the viral proteins PA, PB1, and PB2, is present in every virus particle. If this enzyme were absent from virions, they would never initiate infection, because the (-) strand viral RNAs cannot be translated into protein, and the cell has no enzymes which can copy such long RNA molecules. Of course, additional molecules of the viral RNA polymerase are made in infected cells, but the enzyme that is brought in with the virion is crucial for initiating the infectious cycle.
The influenza viral RNA polymerase is a primer-dependent enzyme. The enzyme cannot copy the (-) strand RNA template without a small piece of RNA that aligns on the template RNA and provides a starting point for RNA synthesis. The primers for influenza viral mRNA synthesis are produced from the cell’s own collection of mRNA molecules. The influenza viral RNA polymerase actually cleaves cell mRNAs near their 5′-ends, generating the primers it requires for RNA synthesis. This process has been called cap snatching, because the primers are made from the 5′-ends of cell mRNAs, which have an unusual chemical structure called a ‘cap’ (box labeled ‘c’ in illustration). Cap-snatching is also a feature of mRNA synthesis of other viruses.
Because RNA-dependent RNA polymerases are not found in mammalian cells, they are an excellent target for inhibition by antiviral compounds. At least one new antiviral drug that targets this enzyme, T-705, is currently in development.

How quickly does this happen? Is it almost instantaneously or a slow process?
Very informative, specially the link to the post on new antiviral drugs. In it, you mention: “DAS181, or Fludase, is unique in that it incorporates a sialidase that removes sialic acids from mucosal membranes, thereby preventing viral attachment via the HA glycoprotein.” and I thought, isn't this bad for the organism?, I mean, sialic acids are there for a reason, right?
How quickly does RNA synthesis occur? Copying of each segment takes
only a few minutes. Once the RNAs are in the nucleus – which takes
minutes also – they are rapidly copied. New virus particles are
produced in the cell within ~4 hr.
The sialidases remove sialic acid from only a small part of the
respiratory tract, just enough to reduce viral entry. They are rapidly
regenerated and thus there is no negative effect on the host.
Certainly if sialic acids were removed permanently from all cells it
would be deterimental.
It takes longer than I thought. Do we know why it takes four hours? Is there a step that takes longer than the rest, that sort of holds it up?
Each step takes time. Virus binding, entry, RNA synthesis, protein
synthesis. Then (which we haven't covered yet) virion assembly and
budding. They all add up. Influenza is actually quite fast. Some
viruses require 24 hr to assemble and exit cells. There isn't any
particular step that is limiting, there are just a lot of them.
Remember the doubling time for a typical cell is 24 hr.
Hey Vincent, thanks! I suppose that's another problem with the virus video, then. It makes it seem so quick, almost instantaneous. and it's not. it's hours long. Is the length of time – about 4 hours – related to how quickly you have take Tamiflu – within two days of symptoms – for it to really help?
Yes, the videos are far too fast, but no one would watch otherwise!
And yes, the replication time is directly related to how much of a
window you have for antivirals to be effective. For flu, that's within
48 hr, because it's a fast (acute) virus. For other, slower viruses,
you may have much more time.
Pingback: The error-prone ways of RNA synthesis
If the (-) strand RNA is also being converted into (+) strand RNA , what system is used for the virus to recognize which strand to take up in the virion? Also (and I know that this goes beyond the scope of this topic) but does the cell have any internal features that allow it to combat viral infections or is its only means of defense to signal the immune system and undergo apoptosis?
The negative strand of RNA must have signals that direct its
incorporation into virions; the plus strand must lack these signals.
However, the specific sequences have not been identified. As for your
defense question – cells have internal defense mechanisms that sense
the presence of foreign RNA and attempt to destroy it. This is carried
out by the innate immune system – which we discussed here:
https://virology.ws/2009/06/03/innate-immune…
The negative strand of RNA must have signals that direct its
incorporation into virions; the plus strand must lack these signals.
However, the specific sequences have not been identified. As for your
defense question – cells have internal defense mechanisms that sense
the presence of foreign RNA and attempt to destroy it. This is carried
out by the innate immune system – which we discussed here:
https://virology.ws/2009/06/03/innate-immune…
I want to know whether there exists a chronological order of influenza protein synthesis?And any paper report?
I’m slightly confused; what is the difference between viras synthesis and viral replication? Does synthesis involve forming new viral proteins, whereas replication involves forming new viral nucleic acids?
Very good question – the answer depends on the virus. Viral RNA
synthesis includes all RNAs made by a virus; mRNAs and genome. So that
term includes viral replication (more accurately, viral genome
replication). For some viruses, such as poliovirus, the genome is the
same as mRNA; so viral RNA synthesis and viral genome replication are
the same. The more generic term ‘viral replication’ is often used to
mean production of new infectious particles.
Thank you so much, this really helped for my powerpoint for AP Bio! I’m just not fully understanding cap snatching though…
Pingback: Viral rna | Moonshotvideo
If the virus doesn’t have a cap already then the viral RNA pol isn’t replicating the – to + then to -, so what is?
The mRNA synthesis and (+)vRNA transcription confuses me. As I understood from this blog, these are two different steps (however, many articles say that mRNA can be used for translation and (-) vRNA formation). So, is the (+) mRNA strand formation catalyzed by viral RNA dependent RNA polymerase or by the host RNA polymerase?
For influenza virus, (+)vRNA and mRNA are different. (+)vRNA cannot be translated into protein. Both RNAs are produced by the viral RNA dependent RNA polymerase, two different forms of the enzyme.
All influenza viral RNA synthesis is carried out by the viral RNA dependent RNA polymerase. The cap plus10-13 nucleotides are derived from host mRNAs.
Looks like you’ve done your research very well.
So a negative sense genome is simply a template for transcription? Or are the nucleotide sequences assembled by addition to the 5′ carbon rather than the 3′?
Thanks!
If the primer for viral mRNA synthesis is the 5′ end derived from host mRNA, then what’s the primer for viral RNA replication?
Hello Professor,
Foe how many hours id mRNA synthesized?
Thank you!
Is the influenza virus considered a provirus?
I have a question do all viruses have both – strand RNA and + strand RNA? If this is the case then do you initially (prior to entry into the cell) only have one kind of RNA?