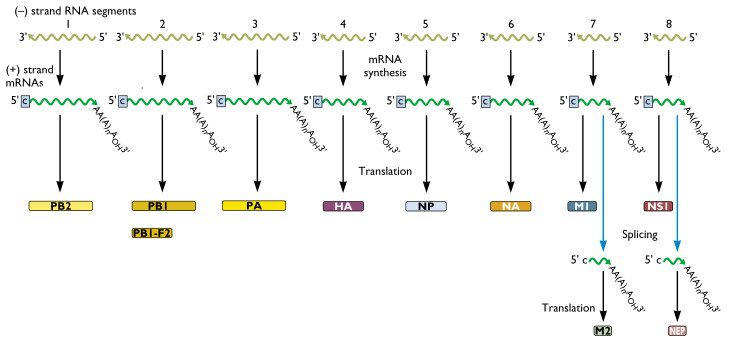

Within the influenza A virion are eight segments of viral RNA. These molecules carry the all the information needed to make new influenza virus particles. These eight RNAs are shown schematically as olive green lines at the top of the illustration. RNAs are chains of four different nucleotides, A, C, G, U. In the case of influenza virus, the eight RNAs are a total of about 14,000 nucleotides in length. The nucleotides make up the genetic code – it is read by the cell’s translation machinery in groups of three, with each triplet specifying an amino acid.
There are two important aspects of these viral RNA that we must consider. First, you can see that the ends of the RNAs are labeled 3′ and 5′. Nucleic acids have polarity, in that one end of the chain is chemically different from the other. Such polarity is represented by 5′ or 3′. The second point is that when a nucleic acid is copied, or duplicated, by enzymes called polymerases, a strand of the complementary polarity is produced. Influenza viral RNAs are called (-), or negative strand RNAs, because they are the opposite polarity of the RNA that is translated to make protein. The RNA molecules that are templates for the synthesis of proteins are defined as having having (+), or positive polarity. Upon entering the cell, the (-) strand influenza viral RNAs must be copied into complementary (+) strands, so that they can serve as templates for proteins. The viral RNAs are copied by an enzyme – called RNA polymerase – that is carried into the cell with the virus.
In the above scheme, the olive green lines are the (-) strand RNAs found in the influenza virion. Once the virion enters the cell, these 8 RNAs are copied into (+) strand mRNAs. Finally, the mRNAs can serve as templates for the synthesis of proteins. The specific viral proteins that are produced by each viral mRNA are shown at the bottom of the illustration. From this picture we see that, for example, RNA segment 4 codes for the viral HA protein, and RNA segment 6 codes for the viral NA protein. Note also that some RNA segments encode for more than one protein. Both influenza A and B viruses have 8 RNA segments, while the influenza C viruses have 7.
Influenza viruses are called (-) strand RNA viruses because of the polarity of the RNA that is carried in the virion. Other RNA viruses – such as poliovirus – are (+) strand RNA viruses, because their genomic RNA can be translated into protein immediately upon entering the cell.
Any questions before we proceed?

This was very good. Great illustration too.
– what does the PB1-F2 tag represent?
Awesome. I'm with you so far. One question maybe for later: I never heard of influenza C viruses. What organisms do these infect? Thanks.
Great illustration.
What does the PB1-F2 tag represent? Another protein from the PB1 strand?
I'm with you so far….
Matt
Thanks a lot for doing this, and for the blog – it's been more interesting, informative and educational than most of the mainstream media coverage so far.
Anything you'd recommend as background reading for the 'educated layperson'?
This is awakening old memories of lectures once attended. So far I'm astounding myself by following completely. I'm looking forward to the next installment.
Thank you for helping to educate non virologists like myself.
It seems to me there needs to be further nomenclature/classification modification. The A/H1N1 classification of this 2009 version does not differentiate clearly from the 1918 virus or the swine/human H1N1 viruses that have been identified before. The concern is that this is a 'new' cross over virus that is human transmissible , where we do not yet know what will be the epedemiology or morbidity/mortaility rates. The nomenclature should reflect the sequence differences that make it 'new'. Some of the comments on phylogeny by the CDC relative to what has been said about the deposited sequences are a bit confusing.
Could you please summarize the differences? A graphical mutation table showing the genes (seqments) compared to related viruses would be most helpful. Are the Polymerase genes the old 'avian' link? What is unique about the H1 subtype that 'puts it in a lonely branch'?
Thank you so much for making this information available to all of us who want to know more about viral infections and influenza. I was going to look for my old virology and microbiology books, but this is more fun!
Thanks.
I just wanted to point out that viral resistance to antiviral drugs often occur at the step of the rna polymerase that copies the (-) strand into the (+) strand. The rna polymerase of the virus is not very faithful and doesn't always copy the (-) strand exactly. Most of the viral rna polymerases are no where near as acurate as our cellular machinery. Thus you can have variations on the virus that can evaid antiviral drugs. This is what you would see with the amatidines and the NA protien. You get a nonexact copy of the NA protien and suddenly amantidine no longer works. This is the variation that causes CDC and WHO big head-aches. The most interesting thing about the inaccuracy of the polymerase is the fact that viruses never evolved a more conservative polymerase that is more accurate. This is one of the great strengths of the viral model, frequent variation. It's also a weakness because the virus can mutate in a direction that makes it less pathogenic. Overall, I would say its a strength.
I am also interested in understanding about -ve sense RNA virus replication: how the -ve sense viral genome replicates and assembles into a mature virion?
Potential Explanations for A H1N1 Flu Mysteries
I put forward the following hypotheses and would like to see WHO to conduct virology experiments and epidemiological studies/investigations to test them:
1. Higher mortality in Mexico is due to:
a. A H1N1 virus could be airborne (not only transmitted by droplets from infected hosts). Therefore, there might be a dose-response relationship. Mexican patients have been exposed to higher virus concentrations with much longer exposure periods than foreign visitors
b. There may be many unidentified infection cases in Mexico, rending an underestimated denominator for mortality calculation. With more and more fatal cases being reported in other countries, differences in mortalities will eventually be reduced
c. Mexico patients may be different from infected visitors in
(1) previous infection(s);
(2) diet/nutrition;
(3) vaccination/immunity/DNA/race;
(4) number of days to seek medical services after developing ILI symptoms;
(5) affordability/availability of medical services;
(6) quality/availability of emergency care;
(7) treatments given;
(8) distribution of patients’ age; density of population/patients;
(9) and so on
d. Seasonal flu was involved in the outbreak, which resulted in a considerable number of cross infections in healthcare settings in Mexico, for medical professionals were/are unable to differentiate A H1N1 infected patients from common flu visitors and isolate “swine flu†victims
e. Climate in Mexico is quite different. Tropical weather is favourable for the breeding and spread of the virus (30oC-35oC may be the optimal growth temperature for A H1N1? Could lower humidity promote infection & spread?). The virulent may also be associated with the environment. Therefore, the prediction from World Meteorology Organization (WMO) and WHO that pandemic will mitigate during the summer might not be true. Actually, pandemic in 1918 and 1968 had peaks in summer. In the tropics, seasonal flu peaks usually occur during hot and rainy months. Meanwhile, the cooling southern hemisphere could be another pool for the pandemic
f. The virulence of A H1N1 might be mitigated while crossing borders because the virus tends to shift/mutate to achieve a balance between killing the hosts and keeping them transmittable
2. Higher fatality between age 20 to 50 because:
a. Many elder people had previously infected by H1N1 viruses and thus are less susceptible than young adults
b. The physiological/metabolic status in 20s and 30s are particular. The immune system of those people in the 20 and 50 age range is prone to be over-activated upon attacked by the virus. A crashed immune system will increase the chance of complications and death
c. People between 20-50 years of age have different occupational and social behavioural factors such as contacts, activities, lifestyle, stress, and so on
Taubenberger and Morens note a similar phenomenon of a “W” shaped mortality histogram and discuss some similar hypothesis with regards to the 1918-19 pandemic.
http://www.cdc.gov/ncidod/eid/vol12no01/05-0979…
article
Influenza C viruses infect only people, and cause mild influenza, with
no antigenic variation.
PB1-F2 is a second protein made from the same RNA that produces PB1.
My textbook, “Principles of Virology”, ASM Press. Start with volume I.
That is where these illustrations come from. If it's too expensive,
try “Principles of Molecular Virology” by AJ Cann.
Not being a scientist, Steven, I assume some of your points could be true. I assume this strain was “churning and burning” around from a few clumps in Mexico, and spread down there much more widely than we know. For a number of economic and social reasons, sentinel detection of these cases was not important. But once the battle flags were risen, a good deal of the rest of the world went into hyper-vigilance, hence the early infection cases from all over the world are now daily news fodder.
The denominator is everything in measuring what the number of deaths means. We read that the sequences of isolates from Mexico have been provided to CDC, and are virtually identical to those CDC has posted, so genetic differences seem not to be an issue.
Thanks for your blog/Twitv's/podcasts.
This is what I understand you to say:
Big Picture:
The main technical issue is to determine if the Mexico strains of A(H1N1) are the same as the other one's studied. For some reason the government of Mexico has not let the public know yet. This could affect innoculation strategy, and probably require to increase to 6, the pharmacological agents at a faster pace. Specifically genetic drift can happen–meaning new higher death viruses can appear given the rapid invasion of pathologies in various regions of the world by this virus.
Medium level picture:
No reason to suspect there will be a spike of total influenza deaths this season in the northern hemisphere, 0.015% of total population in the US, for example (45k). Need to know what will happen in the southern hemisphere, now that winter is setting in. In the main, fast information (maybe not complete) and coordination by WHO, CDC, Canadians and emerging countries are evident world-wide.
My question:
Knowing that the Mexico/2009 InfluenzaA(H1N1) is part of a type of viruses that are unusual in that they copy RNA to DNA in the nucleus of a protein, where one of the segments acts as fusion (HA), and another (NA) aids in transportation to other proteins, isn't it possible that this specific one (the Mexico/2009 ) may teach us that there are heretofore unrecognized innovative strategies in transcription, translation and replication of Influenza viruses?
More specifically:
Can any of the 8 segments be affected either in form or in substance given distinct Ph factors in the specific host protein? Can these Ph factors, in case they are theoretically significant, be adequately measured? Are they clinically relevant?
Pingback: Poliovirus @ Operation Willi
Pingback: How influenza virus inhibits early antiviral responses
Great info.!
Pingback: This Week In Virology « Influenza A (H1N1) Blog
Pingback: This Week In Virology – Esta semana en virologÃa « Gripe por A (H1N1) Blog
Is reverse transcriptase involved in Flu virus life cycle? Do they Flu (-) RNA go to DNA stem before transcribed into (+)RNA?
Reverse transcriptase is not involved in influenza virus replication. The (-) strand influenza viral RNAs are copied to (+) strand RNAs and back to (-) strands by the viral RNA-dependent RNA polymerase.
is the sequences at the end of one segment the same with the sequences at the end of other segmént?
They are highly conserved but not identical.
Does the influenza RDRP proceed 5' to 3' or 3' to 5'? Can the answer be generalized to all viral RDRPs?
All nucleic acid polymerases copy the template from 3' to 5', and
synthesize the product from 5' to 3'.
Hi Prof,
I'm taking a Viology course. I just found your website and I'm happy I did. You're very helpful, so thanks a lot.
One question though, I learnt in class today that Influenza C infects humans and pigs, Influenza B infects only humans and Influenza A infects birds and humans.
Would you minds clarifying please. Thank You very much.
Hi Prof,
I'm taking a Viology course. I just found your website and I'm happy I did. You're very helpful, so thanks a lot.
One question though, I learnt in class today that Influenza C infects humans and pigs, Influenza B infects only humans and Influenza A infects birds and humans.
Would you minds clarifying please. Thank You very much.
can i have please the refrece of the artcile showing this photo (by Vincent Racaniello on 1 May 2009)
Is the life cycle just about the process of it infecting the host? And how the infection debvelops etc or is it about strains of flu and how they run in cycles every so many years?
printed balloons
It should be noted that there is a convention to how the RNA segments are numbered with 1 being the longest segment and 8 being the shortest. So, for flu A, the PB2 gene, #1, is the longest and the NS gene, #8, is the shortest.
What would be different in a virus with DNA vs. RNA?
Think of influenza as a deck of cards, influenza has 14 genes, antigenic drift is when there is a gradual change in the genome and the flu shot from the previous year is partially effective; antigenic shift is when the deck is completely reshuffled, with parts of different flu viruses, avian H1N1, swine flu, recombine, the vaccine is totally ineffective, there is a completely new virus, and worldwide pandemics can occur. Part of why this can happen is that in China, birds are housed above pigs, making the perfect environment for avian and swine viruses combining, much like what happened in Guangdong province in China with the coronavirus SARS developing in a “wet market” for wild animals sold for food being housed next to each other.
 Part of why extremely pathogenic viruses like Ebola have short-lived epidemics is that they kill off their host quickly, and cannot survive and replicate.  Less pathogenic viruses persist because they evolve over time to be less pathogenic, allowing them to continually transmit to hosts and not die out.
 Part of why yellow fever is such a problem is that there is antibody dependent enhancement of immune reaction to the virus, so those that have been infected before or infants with circulating maternal antibodies have more severe immune and hemorrhagic reactions to the yellow fever virus if they are reinfected by an infected mosquito bite. Yellow fever used to have outbreaks as far north as Philadelphia, Pennsylvania and insecticides. Spread of viruses (arboviruses) carried in mosquitoes is aided by shipping old tires that have water carrying the mosquito larvae. Also DDT is extremely effective against the mosquitoes, so disuse has increased mosquito populations. Part of why West Nile virus has an increase in the fall is that the mosquitoes feed on warm-blooded migrating birds and when they leave the area, the mosquitoes feed on humans, and the infection rate increases.
How often are flu immunizations ineffective, because of incorrect anticipation of actual strain?
Does the lead time for production of the flu immunization system ever result in ineffective vaccines because there is enough time for antigen drift?
Does the large population in countries like China, India, or even US result in different population centers (cities) having different flu strains, because the populations are large enough to support more mutations/antigen drift, because of the greater amount of individual replication? (Parallel cases of replication)
Is it know which RNA viral segment creates the hemagglutin portion of the virus ?